Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
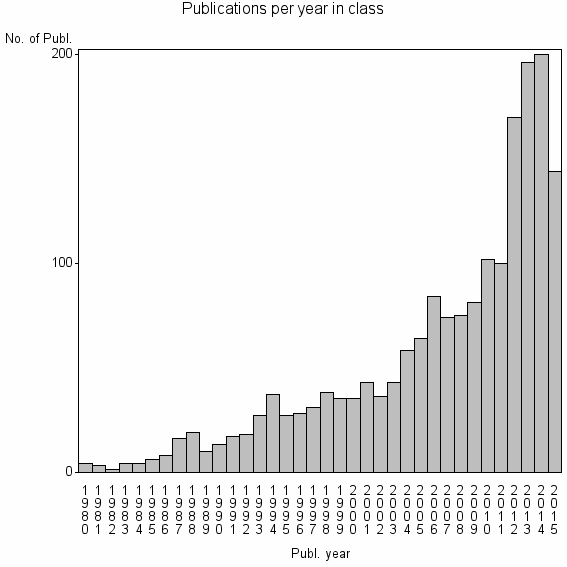
| 3691 | 1851 | 40.1 | 77% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 201 | 20439 | MELAS//MITOCHONDRIAL DNA//LEBERS HEREDITARY OPTIC NEUROPATHY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MITOCHONDRIAL DNA | Journal | 299 | 53% | 21% | 397 |
| 2 | MITOCHONDRIAL GENOME | Author keyword | 257 | 43% | 25% | 454 |
| 3 | MITOGENOME | Author keyword | 170 | 71% | 8% | 139 |
| 4 | COMPLETE MITOCHONDRIAL GENOME | Author keyword | 69 | 56% | 5% | 84 |
| 5 | A PLUS T RICH REGION | Author keyword | 50 | 88% | 1% | 23 |
| 6 | CONTROL REGION | Author keyword | 27 | 18% | 7% | 135 |
| 7 | COMPLETE MTDNA SEQUENCE | Author keyword | 24 | 82% | 1% | 14 |
| 8 | COMPLETE MITOGENOME | Author keyword | 23 | 100% | 1% | 10 |
| 9 | TRNA LIKE SEQUENCE | Author keyword | 21 | 90% | 0% | 9 |
| 10 | MITOGENOMICS | Author keyword | 18 | 45% | 2% | 31 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MITOCHONDRIAL GENOME | 257 | 43% | 25% | 454 | Search MITOCHONDRIAL+GENOME | Search MITOCHONDRIAL+GENOME |
| 2 | MITOGENOME | 170 | 71% | 8% | 139 | Search MITOGENOME | Search MITOGENOME |
| 3 | COMPLETE MITOCHONDRIAL GENOME | 69 | 56% | 5% | 84 | Search COMPLETE+MITOCHONDRIAL+GENOME | Search COMPLETE+MITOCHONDRIAL+GENOME |
| 4 | A PLUS T RICH REGION | 50 | 88% | 1% | 23 | Search A+PLUS+T+RICH+REGION | Search A+PLUS+T+RICH+REGION |
| 5 | CONTROL REGION | 27 | 18% | 7% | 135 | Search CONTROL+REGION | Search CONTROL+REGION |
| 6 | COMPLETE MTDNA SEQUENCE | 24 | 82% | 1% | 14 | Search COMPLETE+MTDNA+SEQUENCE | Search COMPLETE+MTDNA+SEQUENCE |
| 7 | COMPLETE MITOGENOME | 23 | 100% | 1% | 10 | Search COMPLETE+MITOGENOME | Search COMPLETE+MITOGENOME |
| 8 | TRNA LIKE SEQUENCE | 21 | 90% | 0% | 9 | Search TRNA+LIKE+SEQUENCE | Search TRNA+LIKE+SEQUENCE |
| 9 | MITOGENOMICS | 18 | 45% | 2% | 31 | Search MITOGENOMICS | Search MITOGENOMICS |
| 10 | COMPLETE MITOCHONDRIAL DNA SEQUENCE | 18 | 83% | 1% | 10 | Search COMPLETE+MITOCHONDRIAL+DNA+SEQUENCE | Search COMPLETE+MITOCHONDRIAL+DNA+SEQUENCE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | AT RICH REGION | 90 | 88% | 2% | 43 |
| 2 | TRANSFER RNA GENES | 54 | 20% | 13% | 241 |
| 3 | MITOGENOME | 34 | 93% | 1% | 13 |
| 4 | GENE ORGANIZATION | 32 | 20% | 8% | 141 |
| 5 | HIGHER TELEOSTEAN PHYLOGENIES | 31 | 56% | 2% | 38 |
| 6 | MINNOW CYPRINELLA SPILOPTERA | 31 | 92% | 1% | 12 |
| 7 | COMPLETE MTDNA | 30 | 100% | 1% | 12 |
| 8 | DOMESTICATED SILKMOTH | 30 | 84% | 1% | 16 |
| 9 | JAPONICUS CRUSTACEA | 27 | 63% | 1% | 27 |
| 10 | TRUNCATED TRANSFER RNAS | 26 | 100% | 1% | 11 |
Journals |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MITOCHONDRIAL DNA | 299 | 53% | 21% | 397 |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Insect Mitochondrial Genomics: Implications for Evolution and Phylogeny | 2014 | 35 | 129 | 65% |
| Animal mitochondrial genomes | 1999 | 973 | 135 | 67% |
| Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species | 2008 | 146 | 118 | 52% |
| EVOLUTION, WEIGHTING, AND PHYLOGENETIC UTILITY OF MITOCHONDRIAL GENE-SEQUENCES AND A COMPILATION OF CONSERVED POLYMERASE CHAIN-REACTION PRIMERS | 1994 | 2961 | 191 | 20% |
| The complete mitochondrial genome of the Chinese oak silkmoth, Antheraea pernyi (Lepidoptera : Saturniidae) | 2008 | 32 | 33 | 88% |
| Mitogenomic evolution and interrelationships of the cypriniformes (Actinopterygii : Ostariophysi): The first evidence toward resolution of higher-level relationships of the world's largest freshwater fish clade based on 59 whole mitogenome sequences | 2006 | 154 | 79 | 32% |
| Mitochondrial genome data alone are not enough to unambiguously resolve the relationships of Entognatha, Insecta and Crustacea sensu lato (Arthropoda) | 2004 | 85 | 92 | 54% |
| Complete mitochondrial genome of the stonefly Cryptoperla stilifera Sivec (Plecoptera: Peltoperlidae) and the phylogeny of Polyneopteran insects | 2014 | 1 | 25 | 52% |
| Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome 'morphology' | 2002 | 62 | 50 | 54% |
| The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods | 2009 | 18 | 95 | 57% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MARINE LIVING OURCES MOL ENGN | 13 | 36% | 1.5% | 28 |
| 2 | PROV CONSERVAT EXPLOITAT BIOL OU | 8 | 48% | 0.6% | 12 |
| 3 | PHYLOGEN EVOLUT | 7 | 50% | 0.5% | 10 |
| 4 | MARINE BIO ERS ECT SOUTH CHINA SEA | 6 | 100% | 0.2% | 4 |
| 5 | EAST CHINA SEA OCEAN FISHERY OURCES | 4 | 24% | 0.8% | 14 |
| 6 | SPECIAL AQUACULTURE SOURCE | 3 | 100% | 0.2% | 3 |
| 7 | PLANT ENVIRONM PROTECT | 3 | 12% | 1.3% | 24 |
| 8 | PHYLOGENOM EVOLUT | 3 | 45% | 0.3% | 5 |
| 9 | MARINE BIO OURCE SUSTAINABLE UTILIZAT | 3 | 23% | 0.6% | 11 |
| 10 | MOL EVOLUT BIO ERS | 3 | 24% | 0.5% | 10 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000162740 | MITOCHONDRIAL NUCLEOIDS//TFAM//MITOCHONDRIAL NUCLEOID |
| 2 | 0.0000156728 | DOUBLY UNIPARENTAL INHERITANCE//PATERNAL LEAKAGE//MITOCHONDRIAL POLARITY |
| 3 | 0.0000107923 | NUMT//NUMTS//NUMTDNA |
| 4 | 0.0000086337 | TETRAODONTIFORMES//MOLIDAE//RANZANIA LAEVIS |
| 5 | 0.0000076392 | STANDARDS IN GENOMIC SCIENCES//BIOL DATA MANAGEMENT TECHNOL//GEBA |
| 6 | 0.0000075554 | TRNA NUCLEOTIDYLTRANSFERASE//CCA ADDING ENZYME//TRNAHIS GUANYLYLTRANSFERASE |
| 7 | 0.0000064784 | ONYCHOPHORA//PYCNOGONIDA//PERIPATOPSIDAE |
| 8 | 0.0000062616 | OPSARIICHTHYS//NESTED CLADE ANALYSIS//POPULATION EXPANSION |
| 9 | 0.0000057241 | CODON MODEL//HETEROTACHY//PHYLOGENETIC INVARIANTS |
| 10 | 0.0000053097 | MUSEUM NAT SCI//SPECIES LIMITS//BELL MUSEUM |