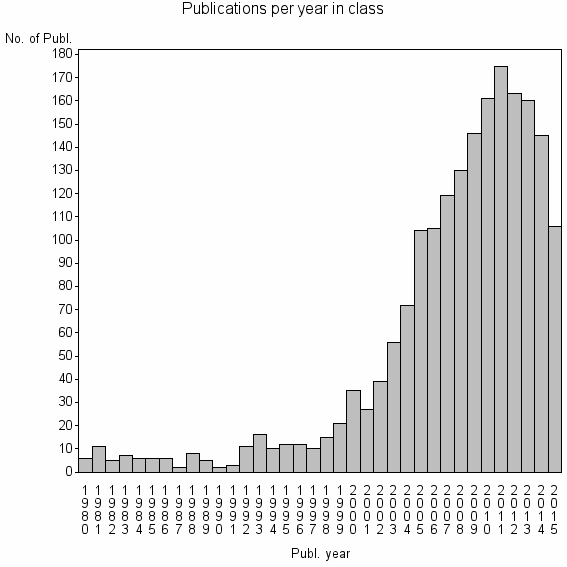
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 3401 | 1917 | 48.5 | 77% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PSEUDO AMINO ACID COMPOSITION | Author keyword | 284 | 91% | 6% | 116 |
| 2 | COMPUTAT MUTAT PROJECT | Address | 82 | 92% | 2% | 33 |
| 3 | JACKKNIFE TEST | Author keyword | 61 | 86% | 2% | 31 |
| 4 | PROTEIN STRUCTURAL CLASS | Author keyword | 56 | 80% | 2% | 35 |
| 5 | CHOUS PSEUDO AMINO ACID COMPOSITION | Author keyword | 41 | 100% | 1% | 15 |
| 6 | COVARIANT DISCRIMINANT ALGORITHM | Author keyword | 38 | 89% | 1% | 17 |
| 7 | FUNCTIONAL DOMAIN COMPOSITION | Author keyword | 38 | 93% | 1% | 14 |
| 8 | AMPHIPHILIC PSEUDO AMINO ACID COMPOSITION | Author keyword | 31 | 92% | 1% | 12 |
| 9 | DIAMOND OPERATOR | Author keyword | 31 | 92% | 1% | 12 |
| 10 | PROTEIN SUBCELLULAR LOCALIZATION | Author keyword | 28 | 61% | 2% | 30 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | FUNCTIONAL DOMAIN COMPOSITION | 457 | 88% | 11% | 217 |
| 2 | ENZYME SUBFAMILY CLASSES | 422 | 97% | 6% | 115 |
| 3 | STRUCTURAL CLASS PREDICTION | 389 | 90% | 9% | 167 |
| 4 | SUBCELLULAR LOCATION PREDICTION | 363 | 89% | 9% | 166 |
| 5 | ENSEMBLE CLASSIFIER | 362 | 92% | 8% | 145 |
| 6 | AMINO ACID COMPOSITION | 299 | 34% | 37% | 718 |
| 7 | STRUCTURAL CLASSES | 219 | 83% | 6% | 123 |
| 8 | IMPROVED HYBRID APPROACH | 197 | 98% | 3% | 50 |
| 9 | MODIFIED MAHALANOBIS DISCRIMINANT | 171 | 96% | 3% | 52 |
| 10 | LOCATION PREDICTION | 166 | 87% | 4% | 81 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Some remarks on protein attribute prediction and pseudo amino acid composition | 2011 | 329 | 197 | 89% |
| iRSpot-TNCPseAAC: Identify Recombination Spots with Trinucleotide Composition and Pseudo Amino Acid Components | 2014 | 48 | 110 | 88% |
| Some remarks on predicting multi-label attributes in molecular biosystems | 2013 | 66 | 58 | 90% |
| Locating proteins in the cell using TargetP, SignalP and related tools | 2007 | 1418 | 110 | 38% |
| Recent progress in protein subcellular location prediction | 2007 | 474 | 77 | 91% |
| Pseudo Amino Acid Composition and its Applications in Bioinformatics, Proteomics and System Biology | 2009 | 137 | 175 | 84% |
| iNR-Drug: Predicting the Interaction of Drugs with Nuclear Receptors in Cellular Networking | 2014 | 22 | 156 | 85% |
| GPCR-CA: A Cellular Automaton Image Approach for Predicting G-Protein-Coupled Receptor Functional Classes | 2009 | 129 | 96 | 83% |
| Graphic Rule for Drug Metabolism Systems | 2010 | 94 | 50 | 68% |
| Prediction of protein structural classes by Chou's pseudo amino acid composition: approached using continuous wavelet transform and principal component analysis | 2009 | 54 | 107 | 85% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | COMPUTAT MUTAT PROJECT | 82 | 92% | 1.7% | 33 |
| 2 | NEUROINFORMATMINIST EDUC | 15 | 56% | 0.9% | 18 |
| 3 | COMP AIDED DRUG DISCOVERY | 13 | 35% | 1.6% | 30 |
| 4 | ENGN NONFOOD BIOREFINERY | 10 | 32% | 1.3% | 25 |
| 5 | STATE NONFOOD BIOMASS ENZYME TECHNOL | 9 | 44% | 0.8% | 16 |
| 6 | THEORET BIOPHYS | 8 | 24% | 1.5% | 28 |
| 7 | GUANGXI BIOREFINERY | 8 | 42% | 0.7% | 14 |
| 8 | UPJOHN S | 8 | 50% | 0.6% | 11 |
| 9 | GENOM COMPUTAT BIOL | 7 | 23% | 1.5% | 28 |
| 10 | BIOINFORMAT DRUG DISCOVERY | 7 | 57% | 0.4% | 8 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000157726 | SECONDARY STRUCTURE PREDICTION//PROTEIN SECONDARY STRUCTURE PREDICTION//COMPOUND PYRAMID MODEL |
| 2 | 0.0000132699 | BIOINFORMAT TELEMED//HYDROPHOBICITY DEFICIENCY//HELIX INTERACTIONS |
| 3 | 0.0000127909 | TOMOCOMD CARDD SOFTWARE//CHEM BIOACT//UNIDAD INVEST DISENO FARMACOS CONECTIVIDAD MOL |
| 4 | 0.0000120154 | INTERACTION PROPENSITY//RNA BINDING RESIDUES//COMPUTAT INTELLIGENCE LEARNING DISCOVERY |
| 5 | 0.0000104356 | GRAPHICAL REPRESENTATION//ALIGNMENT FREE//SIMILARITIES DISSIMILARITIES |
| 6 | 0.0000065997 | EMBL OUTSTN//PROT INFORMAT OURCE//HLTH SCI TECHNOL ICIR |
| 7 | 0.0000064477 | PROTEIN STRUCTURE PREDICTION//FOLD RECOGNITION//CASP |
| 8 | 0.0000059107 | COMPLEX SCI SOFTWARE//3D ZERNIKE DESCRIPTOR//PROTEIN SURFACE SHAPE |
| 9 | 0.0000048915 | TRANSMEMBRANE HELIX//MEMBRANE PROTEIN FOLDING//TRANSMEMBRANE HELICES |
| 10 | 0.0000047201 | ELECTRON ION INTERACTION POTENTIAL//INFORMATIONAL SPECTRUM METHOD//DECEPTIVE IMPRINTING |