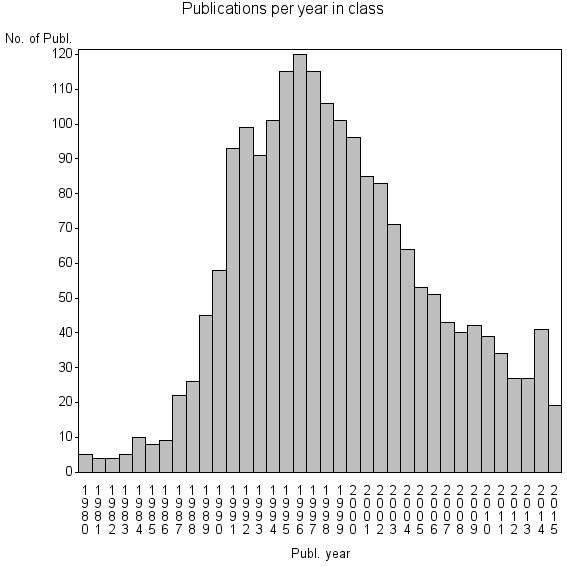
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 3254 | 1952 | 44.7 | 88% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1002 | 9892 | PHOSPHOLIPASE D//DIACYLGLYCEROL KINASE//PROTEIN KINASE C |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PHOSPHOLIPASE D | Author keyword | 330 | 44% | 29% | 569 |
| 2 | PHOSPHATIDIC ACID | Author keyword | 49 | 25% | 9% | 169 |
| 3 | PHOSPHOLIPASE D2 | Author keyword | 40 | 82% | 1% | 23 |
| 4 | PHOSPHOLIPASE D1 | Author keyword | 24 | 61% | 1% | 25 |
| 5 | PHOSPHATIDYLETHANOL | Author keyword | 22 | 42% | 2% | 40 |
| 6 | PLD1 | Author keyword | 18 | 67% | 1% | 16 |
| 7 | PLD2 | Author keyword | 17 | 72% | 1% | 13 |
| 8 | PHOSPHOLIPASE D PLD | Author keyword | 14 | 55% | 1% | 18 |
| 9 | PHOSPHATIDYLBUTANOL | Author keyword | 8 | 70% | 0% | 7 |
| 10 | ADP RIBOSYLATION FACTOR | Author keyword | 7 | 19% | 2% | 32 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PHOSPHOLIPASE D | 330 | 44% | 29% | 569 | Search PHOSPHOLIPASE+D | Search PHOSPHOLIPASE+D |
| 2 | PHOSPHATIDIC ACID | 49 | 25% | 9% | 169 | Search PHOSPHATIDIC+ACID | Search PHOSPHATIDIC+ACID |
| 3 | PHOSPHOLIPASE D2 | 40 | 82% | 1% | 23 | Search PHOSPHOLIPASE+D2 | Search PHOSPHOLIPASE+D2 |
| 4 | PHOSPHOLIPASE D1 | 24 | 61% | 1% | 25 | Search PHOSPHOLIPASE+D1 | Search PHOSPHOLIPASE+D1 |
| 5 | PHOSPHATIDYLETHANOL | 22 | 42% | 2% | 40 | Search PHOSPHATIDYLETHANOL | Search PHOSPHATIDYLETHANOL |
| 6 | PLD1 | 18 | 67% | 1% | 16 | Search PLD1 | Search PLD1 |
| 7 | PLD2 | 17 | 72% | 1% | 13 | Search PLD2 | Search PLD2 |
| 8 | PHOSPHOLIPASE D PLD | 14 | 55% | 1% | 18 | Search PHOSPHOLIPASE+D+PLD | Search PHOSPHOLIPASE+D+PLD |
| 9 | PHOSPHATIDYLBUTANOL | 8 | 70% | 0% | 7 | Search PHOSPHATIDYLBUTANOL | Search PHOSPHATIDYLBUTANOL |
| 10 | ADP RIBOSYLATION FACTOR | 7 | 19% | 2% | 32 | Search ADP+RIBOSYLATION+FACTOR | Search ADP+RIBOSYLATION+FACTOR |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | D ACTIVATION | 193 | 68% | 9% | 171 |
| 2 | ADP RIBOSYLATION FACTOR | 154 | 30% | 22% | 428 |
| 3 | HL 60 GRANULOCYTES | 141 | 73% | 6% | 108 |
| 4 | PHOSPHATIDYLCHOLINE HYDROLYSIS | 139 | 55% | 9% | 174 |
| 5 | PHOSPHATIDIC ACID | 120 | 22% | 25% | 481 |
| 6 | D DEFINES | 52 | 68% | 2% | 45 |
| 7 | PHOSPHATIDYLCHOLINE BREAKDOWN | 41 | 45% | 4% | 69 |
| 8 | D ISOZYMES | 38 | 93% | 1% | 14 |
| 9 | D MEDIATED HYDROLYSIS | 31 | 82% | 1% | 18 |
| 10 | D PLD ISOZYMES | 24 | 91% | 1% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Phospholipase D: a lipid centric review | 2005 | 246 | 157 | 75% |
| Phospholipase D: molecular and cell biology of a novel gene family | 2000 | 378 | 220 | 64% |
| Phospholipase D | 2004 | 221 | 313 | 66% |
| The phospholipase D superfamily as therapeutic targets | 2015 | 1 | 55 | 58% |
| Phospholipase D in cell proliferation and cancer | 2003 | 175 | 155 | 69% |
| Regulation of phospholipase D | 1999 | 287 | 144 | 74% |
| Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners | 2012 | 30 | 121 | 64% |
| Molecular mechanisms of PLD function in membrane traffic | 2008 | 65 | 75 | 61% |
| New developments in phospholipase D | 1997 | 272 | 73 | 79% |
| PHOSPHATIDYLCHOLINE BREAKDOWN AND SIGNAL-TRANSDUCTION | 1994 | 814 | 249 | 43% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOMED OURCES | 2 | 15% | 0.7% | 13 |
| 2 | MOL CELLULAR BIOL PATHOBIOL PROGRAM | 1 | 38% | 0.2% | 3 |
| 3 | EXPT CELL | 1 | 100% | 0.1% | 2 |
| 4 | HUMAN ECOL HUMAN ANAT CELL SCI | 1 | 100% | 0.1% | 2 |
| 5 | MOL MED 438 | 1 | 50% | 0.1% | 2 |
| 6 | CELL SCI PHYSIOL | 1 | 33% | 0.2% | 3 |
| 7 | ENVIRONM CELL PONSES | 1 | 40% | 0.1% | 2 |
| 8 | SOCHO GU | 1 | 12% | 0.4% | 8 |
| 9 | SIGNALLING | 1 | 14% | 0.3% | 6 |
| 10 | CARDIAC MEMBRANE BIOL | 1 | 33% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000140000 | DIACYLGLYCEROL KINASE//DIACYLGLYCEROL KINASE DGK//DGK ZETA |
| 2 | 0.0000101652 | ADP RIBOSYLATION FACTOR//ARF6//CYTOHESIN |
| 3 | 0.0000092475 | SECT PLANT PHYSIOL//PHOSPHATIDIC ACID//PHOSPHOLIPID SIGNALLING |
| 4 | 0.0000076503 | TRANSPHOSPHATIDYLATION//STRUCTURED PHOSPHOLIPIDS//ENZYMATIC DEGUMMING |
| 5 | 0.0000060681 | CYTIDYLYLTRANSFERASE//CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE//CTP PHOSPHOCHOLINE CYTIDYLYLTRANSFERASE |
| 6 | 0.0000057051 | TMEM132A//C FOS SERUM RESPONSE ELEMENT//FAS FASL INTERACTIONS |
| 7 | 0.0000055861 | PHOSPHATIDATE PHOSPHOHYDROLASE//SN GLYCEROL 3 PHOSPHATE//GLYCEROPHOSPHATE ACYLTRANSFERASE |
| 8 | 0.0000055766 | LYSOPHOSPHATIDIC ACID//AUTOTAXIN//LYSOPHOSPHOLIPASE D |
| 9 | 0.0000055257 | PHOSPHOLIPASE C DELTA 1//PHOSPHOLIPASE C GAMMA 1//GENOME BIOSIGNAL |
| 10 | 0.0000052077 | DIACYLGLYCEROL FORMATION//INOSITOL 1 3 4 5 TETRAKISPHOSPHATE//BRAIN CORTICAL SLICES |