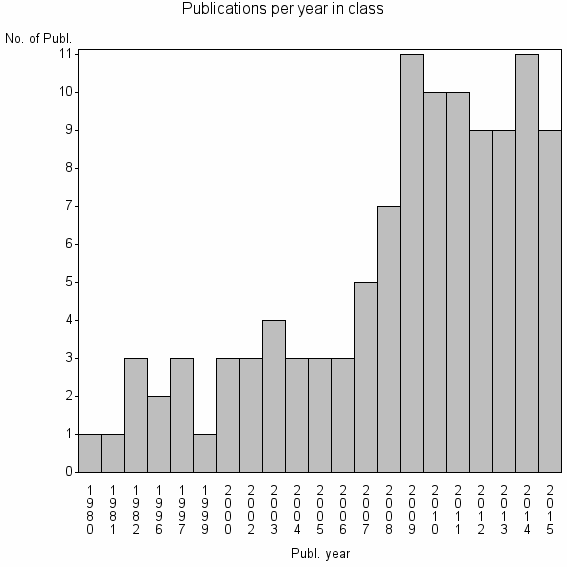
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 32245 | 111 | 50.5 | 86% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | LECTIN RECEPTOR LIKE KINASE | Author keyword | 11 | 100% | 5% | 6 |
| 2 | SECT MOL BIOTECHNOL | Address | 3 | 50% | 5% | 5 |
| 3 | LECTIN RECEPTOR KINASES | Author keyword | 3 | 100% | 3% | 3 |
| 4 | APYRASES | Author keyword | 1 | 33% | 2% | 2 |
| 5 | LECTIN RECEPTOR KINASE | Author keyword | 1 | 33% | 2% | 2 |
| 6 | SALINITY STRESS TOLERANCE | Author keyword | 1 | 33% | 2% | 2 |
| 7 | CAMP MEDIATED | Author keyword | 1 | 50% | 1% | 1 |
| 8 | MOL CELL BIOL HEBEI PROV | Address | 1 | 50% | 1% | 1 |
| 9 | POPULUS NIGRA VAR ITALICA | Author keyword | 1 | 50% | 1% | 1 |
| 10 | TYR NITRATION | Author keyword | 1 | 50% | 1% | 1 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | LECTIN RECEPTOR LIKE KINASE | 11 | 100% | 5% | 6 | Search LECTIN+RECEPTOR+LIKE+KINASE | Search LECTIN+RECEPTOR+LIKE+KINASE |
| 2 | LECTIN RECEPTOR KINASES | 3 | 100% | 3% | 3 | Search LECTIN+RECEPTOR+KINASES | Search LECTIN+RECEPTOR+KINASES |
| 3 | APYRASES | 1 | 33% | 2% | 2 | Search APYRASES | Search APYRASES |
| 4 | LECTIN RECEPTOR KINASE | 1 | 33% | 2% | 2 | Search LECTIN+RECEPTOR+KINASE | Search LECTIN+RECEPTOR+KINASE |
| 5 | SALINITY STRESS TOLERANCE | 1 | 33% | 2% | 2 | Search SALINITY+STRESS+TOLERANCE | Search SALINITY+STRESS+TOLERANCE |
| 6 | CAMP MEDIATED | 1 | 50% | 1% | 1 | Search CAMP+MEDIATED | Search CAMP+MEDIATED |
| 7 | POPULUS NIGRA VAR ITALICA | 1 | 50% | 1% | 1 | Search POPULUS+NIGRA+VAR+ITALICA | Search POPULUS+NIGRA+VAR+ITALICA |
| 8 | TYR NITRATION | 1 | 50% | 1% | 1 | Search TYR+NITRATION | Search TYR+NITRATION |
| 9 | ANIMAL IMMUNITY | 0 | 33% | 1% | 1 | Search ANIMAL+IMMUNITY | Search ANIMAL+IMMUNITY |
| 10 | EXTRACELLULAR ATP EATP | 0 | 33% | 1% | 1 | Search EXTRACELLULAR+ATP+EATP | Search EXTRACELLULAR+ATP+EATP |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SIGNALING ROLE | 4 | 41% | 6% | 7 |
| 2 | PLANT RECEPTOR | 2 | 67% | 2% | 2 |
| 3 | POLLEN GERMINATION | 2 | 12% | 14% | 16 |
| 4 | APYRASES | 2 | 43% | 3% | 3 |
| 5 | LEGUME ROOTS | 1 | 33% | 2% | 2 |
| 6 | MILTIORRHIZA HAIRY ROOTS | 1 | 50% | 1% | 1 |
| 7 | PROTEIN KINASE TARGET | 1 | 50% | 1% | 1 |
| 8 | SYNAPTOTAGMIN SYT1 | 1 | 50% | 1% | 1 |
| 9 | DASYCLADUS VERMICULARIS | 0 | 20% | 2% | 2 |
| 10 | TISSUE REUNION | 0 | 33% | 1% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Extracellular ATP is a central signaling molecule in plant stress responses | 2014 | 9 | 47 | 64% |
| Lectin receptor kinases in plant innate immunity | 2013 | 15 | 42 | 33% |
| Extracellular ATP signaling in plants | 2010 | 40 | 71 | 41% |
| Extracellular ATP: an unexpected role as a signaler in plants | 2007 | 70 | 26 | 50% |
| Apyrases, extracellular ATP and the regulation of growth | 2011 | 8 | 44 | 68% |
| Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants | 2014 | 5 | 84 | 36% |
| Extracellular ATP, a danger signal, is recognized by DORN1 in Arabidopsis | 2014 | 1 | 73 | 52% |
| Current status and proposed roles for nitric oxide as a key mediator of the effects of extracellular nucleotides on plant growth | 2013 | 4 | 86 | 36% |
| Breakthroughs spotlighting roles for extracellular nucleotides and apyrases in stress responses and growth and development | 2014 | 3 | 69 | 36% |
| Plant extracellular ATP signalling: new insight from proteomics | 2012 | 5 | 75 | 33% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SECT MOL BIOTECHNOL | 3 | 50% | 4.5% | 5 |
| 2 | MOL CELL BIOL HEBEI PROV | 1 | 50% | 0.9% | 1 |
| 3 | PLANT MOL BIOL BIOTECHNOL PMBBRC | 0 | 20% | 1.8% | 2 |
| 4 | BIOL S 311 | 0 | 33% | 0.9% | 1 |
| 5 | BIOL SCI MOL CYTOL CORE | 0 | 33% | 0.9% | 1 |
| 6 | CREAT GENE TECHNOL LTD | 0 | 33% | 0.9% | 1 |
| 7 | SIGNAUX MESSAGES CELLULAI CHEZ VEGETAUX | 0 | 20% | 0.9% | 1 |
| 8 | PLANT MICROBIOL PATHOL | 0 | 14% | 0.9% | 1 |
| 9 | UNIV BIOTECHNOL ASTURIAS | 0 | 14% | 0.9% | 1 |
| 10 | SHANGHAI BIOL SCI SIBS | 0 | 10% | 0.9% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000214514 | NTPDASE//ATP DIPHOSPHOHYDROLASE//ECTO ATPASE |
| 2 | 0.0000127626 | COOPERAT DEMONSTRAT CENTRIFUGE TECH//BECKMAN COULTER LTD CO//GRP SENALIZAC MOL SISTEMAS ANTIOXIDANTES PLANTA |
| 3 | 0.0000126752 | AGB1//HETEROTRIMERIC G PROTEIN//HETEROTRIMERIC G PROTEIN BETA SUBUNIT |
| 4 | 0.0000104000 | CAFFEINE DEGRADATION//METAB BIOL GRPBUNKYO KU//CAFFEINE BIOSYNTHESIS |
| 5 | 0.0000098753 | GLUTATHIONE CONJUGATE PUMP//PLANT BIOL MOL PLANT PHYSIOL//PRIMARY TRANSPORT |
| 6 | 0.0000096554 | TYPE III EFFECTOR//MOLECULAR PLANT-MICROBE INTERACTIONS//SGT1 |
| 7 | 0.0000088856 | POLLEN TUBE//TIP GROWTH//ROP GTPASE |
| 8 | 0.0000086184 | GUARD CELL//GUARD CELLS//SV CHANNEL |
| 9 | 0.0000079474 | LEGUME LECTIN//LECTIN//PLANT LECTIN |
| 10 | 0.0000078535 | ELICITIN//ELICITOR//CRYPTOGEIN |