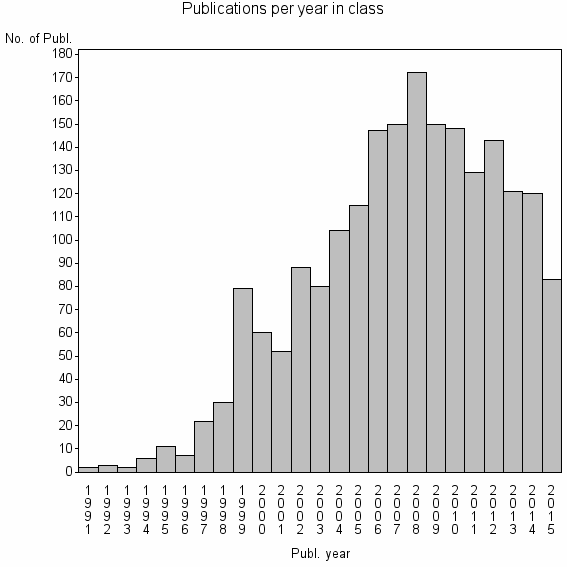
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 2964 | 2024 | 42.6 | 87% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1757 | 5896 | SINGLE MOLECULE FORCE SPECTROSCOPY//FORCE SPECTROSCOPY//MECHANICAL UNFOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | SINGLE MOLECULE FORCE SPECTROSCOPY | Author keyword | 81 | 59% | 4% | 90 |
| 2 | MECHANICAL UNFOLDING | Author keyword | 44 | 76% | 2% | 31 |
| 3 | FORCE SPECTROSCOPY | Author keyword | 44 | 32% | 5% | 111 |
| 4 | LEHRSTUHL ANGEW PHYS | Address | 25 | 37% | 3% | 55 |
| 5 | PROTEIN STRETCHING | Author keyword | 23 | 86% | 1% | 12 |
| 6 | DYNAMIC FORCE SPECTROSCOPY | Author keyword | 18 | 50% | 1% | 26 |
| 7 | PHYS E22 | Address | 14 | 32% | 2% | 37 |
| 8 | STATE PRECIS MEASUREMENTS TECHNOL R | Address | 12 | 86% | 0% | 6 |
| 9 | IMETUM | Address | 9 | 43% | 1% | 17 |
| 10 | UNBINDING FORCE | Author keyword | 9 | 64% | 0% | 9 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | TITIN IMMUNOGLOBULIN DOMAINS | 160 | 73% | 6% | 124 |
| 2 | SINGLE PROTEIN | 94 | 60% | 5% | 103 |
| 3 | ADHESION BONDS | 94 | 75% | 3% | 67 |
| 4 | CLAMP SPECTROSCOPY | 72 | 93% | 1% | 27 |
| 5 | MOLECULE FORCE SPECTROSCOPY | 70 | 44% | 6% | 122 |
| 6 | RECOGNITION EVENTS | 55 | 66% | 3% | 51 |
| 7 | UNFOLDING PATHWAYS | 46 | 51% | 3% | 65 |
| 8 | ANTIGEN BINDING FORCES | 45 | 94% | 1% | 16 |
| 9 | DESORPTION FORCE | 45 | 90% | 1% | 19 |
| 10 | TITIN | 38 | 19% | 9% | 178 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Detection and localization of single molecular recognition events using atomic force microscopy | 2006 | 460 | 74 | 55% |
| Single-molecule experiments in vitro and in silico | 2007 | 272 | 60 | 62% |
| Force and function: probing proteins with AFM-based force spectroscopy | 2009 | 112 | 79 | 66% |
| Atomic force microscopy: A multifaceted tool to study membrane proteins and their interactions with ligands | 2014 | 14 | 118 | 47% |
| Multiparametric imaging of biological systems by force-distance curve-based AFM | 2013 | 51 | 77 | 39% |
| Reconstructing Folding Energy Landscapes by Single-Molecule Force Spectroscopy | 2014 | 14 | 111 | 41% |
| Atomic force microscopy as a multifunctional molecular toolbox in nanobiotechnology | 2008 | 240 | 94 | 37% |
| The structure and function of cell membranes examined by atomic force microscopy and single-molecule force spectroscopy | 2015 | 2 | 153 | 32% |
| Stretching single polysaccharides and proteins using atomic force microscopy | 2012 | 33 | 57 | 63% |
| Force probing surfaces of living cells to molecular resolution | 2009 | 121 | 90 | 41% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | LEHRSTUHL ANGEW PHYS | 25 | 37% | 2.7% | 55 |
| 2 | PHYS E22 | 14 | 32% | 1.8% | 37 |
| 3 | STATE PRECIS MEASUREMENTS TECHNOL R | 12 | 86% | 0.3% | 6 |
| 4 | IMETUM | 9 | 43% | 0.8% | 17 |
| 5 | NANONEUROBIOPHYS GRP | 8 | 75% | 0.3% | 6 |
| 6 | CHRISTIAN DOPPLER NANOSCOP METHODS BIOPHYS | 6 | 42% | 0.5% | 11 |
| 7 | ERNST BERL MAKROMOL CHEM | 6 | 100% | 0.2% | 4 |
| 8 | CHAIR PL PHYS | 5 | 30% | 0.7% | 15 |
| 9 | ATOMIST MOL MECH | 5 | 18% | 1.3% | 26 |
| 10 | SINGLE MOL STUDY | 4 | 38% | 0.4% | 9 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000198078 | BMSE PROGRAM//MAGNETIC TWEEZERS//SMALL BIOSYST |
| 2 | 0.0000182333 | UNITE CHIM INTER ES//SINGLE CELL FORCE SPECTROSCOPY//BACTERIAL INTERACTION FORCES |
| 3 | 0.0000137870 | NANONEWTON//SPRING CONSTANT//NORMAL SPRING CONSTANT |
| 4 | 0.0000130467 | CATCH BONDS//BELL MODEL//CELL ROLLING |
| 5 | 0.0000099971 | DNA NETWORK//FOR UNGSZENTRUM IBI STRUCT BIOL 2//CRYO AFM |
| 6 | 0.0000093527 | ULTRASONIC DEGRADATION//MECHANOPHORES//TRANSIENT ELONGATIONAL FLOW |
| 7 | 0.0000082731 | CELL MECHANICS//MICROPIPETTE ASPIRATION//CELL STIFFNESS |
| 8 | 0.0000068555 | TITIN//CONNECTIN//OBSCURIN |
| 9 | 0.0000064457 | MEAN SQUARED RADIUS OF GYRATION//POWERSPECTRA//SINGLE CHAINS |
| 10 | 0.0000059846 | PROTEIN FOLDING//PHI VALUE//CONTACT ORDER |