Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
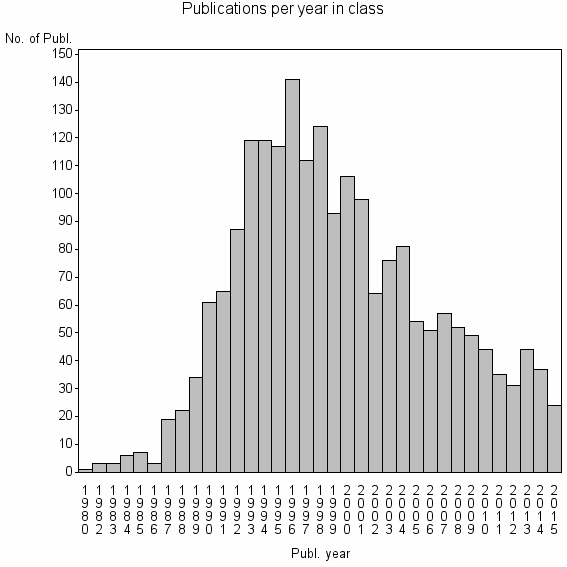
| 2907 | 2039 | 48.6 | 92% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | TFIID | Author keyword | 91 | 66% | 4% | 85 |
| 2 | TFIIA | Author keyword | 34 | 79% | 1% | 22 |
| 3 | MOT1 | Author keyword | 21 | 90% | 0% | 9 |
| 4 | TFIIE | Author keyword | 21 | 85% | 1% | 11 |
| 5 | TFIIB | Author keyword | 20 | 57% | 1% | 24 |
| 6 | TAFS | Author keyword | 18 | 61% | 1% | 19 |
| 7 | GENERAL TRANSCRIPTION FACTOR | Author keyword | 17 | 70% | 1% | 14 |
| 8 | RNA POLYMERASE II | Author keyword | 14 | 13% | 5% | 102 |
| 9 | TBP | Author keyword | 13 | 16% | 4% | 74 |
| 10 | GENERAL TRANSCRIPTION FACTORS | Author keyword | 13 | 58% | 1% | 15 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TFIID | 91 | 66% | 4% | 85 | Search TFIID | Search TFIID |
| 2 | TFIIA | 34 | 79% | 1% | 22 | Search TFIIA | Search TFIIA |
| 3 | MOT1 | 21 | 90% | 0% | 9 | Search MOT1 | Search MOT1 |
| 4 | TFIIE | 21 | 85% | 1% | 11 | Search TFIIE | Search TFIIE |
| 5 | TFIIB | 20 | 57% | 1% | 24 | Search TFIIB | Search TFIIB |
| 6 | TAFS | 18 | 61% | 1% | 19 | Search TAFS | Search TAFS |
| 7 | GENERAL TRANSCRIPTION FACTOR | 17 | 70% | 1% | 14 | Search GENERAL+TRANSCRIPTION+FACTOR | Search GENERAL+TRANSCRIPTION+FACTOR |
| 8 | RNA POLYMERASE II | 14 | 13% | 5% | 102 | Search RNA+POLYMERASE+II | Search RNA+POLYMERASE+II |
| 9 | TBP | 13 | 16% | 4% | 74 | Search TBP | Search TBP |
| 10 | GENERAL TRANSCRIPTION FACTORS | 13 | 58% | 1% | 15 | Search GENERAL+TRANSCRIPTION+FACTORS | Search GENERAL+TRANSCRIPTION+FACTORS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PREINITIATION COMPLEX | 171 | 44% | 14% | 292 |
| 2 | TATA BINDING PROTEIN | 141 | 26% | 23% | 465 |
| 3 | YEAST TFIIA TBP DNA COMPLEX | 119 | 71% | 5% | 95 |
| 4 | RNA POLYMERASE II | 105 | 10% | 48% | 984 |
| 5 | TFIID COMPLEX | 80 | 56% | 5% | 96 |
| 6 | HISTONE LIKE TAFS | 72 | 93% | 1% | 27 |
| 7 | TBP | 60 | 33% | 7% | 148 |
| 8 | TBP BINDING | 53 | 69% | 2% | 46 |
| 9 | START SITE SELECTION | 52 | 51% | 4% | 74 |
| 10 | MINIMAL SET | 52 | 68% | 2% | 45 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The RNA polymerase II core promoter | 2003 | 528 | 220 | 60% |
| Regulation of gene expression via the core promoter and the basal transcriptional machinery | 2010 | 111 | 51 | 76% |
| The general transcription machinery and general cofactors | 2006 | 325 | 747 | 52% |
| The role of general initiation factors in transcription by RNA polymerase II | 1996 | 669 | 62 | 95% |
| Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation | 2010 | 72 | 66 | 80% |
| Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression | 2003 | 155 | 77 | 79% |
| Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes | 1997 | 470 | 97 | 65% |
| Structure and mechanism of the RNA polymerase II transcription machinery | 2004 | 233 | 124 | 60% |
| The general transcription factors of RNA polymerase II | 1996 | 719 | 265 | 65% |
| TRANSCRIPTIONAL ACTIVATION - A COMPLEX PUZZLE WITH FEW EASY PIECES | 1994 | 886 | 22 | 64% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | LI KA SHING BIOMED HLTH SCI | 6 | 48% | 0.5% | 10 |
| 2 | ROGER ADAMS 430 | 6 | 80% | 0.2% | 4 |
| 3 | SECT NMR SPECT | 4 | 75% | 0.1% | 3 |
| 4 | CIRM EXCELLENCE | 3 | 57% | 0.2% | 4 |
| 5 | FYSIOL CHEM | 3 | 100% | 0.1% | 3 |
| 6 | GENE FUNCT ANIM | 3 | 25% | 0.5% | 11 |
| 7 | ADV MOL BIOL IMMUNOL | 3 | 60% | 0.1% | 3 |
| 8 | NUCL ACIDS ENZYMOL | 3 | 35% | 0.3% | 6 |
| 9 | N FREAR 456 | 2 | 67% | 0.1% | 2 |
| 10 | DEV BIOL BUNKYO KU | 2 | 50% | 0.1% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000235142 | MED12//MEDIATOR COMPLEX//MED19 |
| 2 | 0.0000110936 | P TEFB//CDK9//HEXIM1 |
| 3 | 0.0000109344 | USF1//UPSTREAM TRANSCRIPTION FACTOR 1//USF |
| 4 | 0.0000108455 | RNA POLYMERASE III//TFIIIB//SNAPC |
| 5 | 0.0000103986 | MYST//MOZ//HISTONE ACETYLTRANSFERASE |
| 6 | 0.0000089999 | RPB7//RPB4//INTERSPECIFIC COMPLEMENTATION |
| 7 | 0.0000087413 | GAL4P//GAL80//GAL GENES |
| 8 | 0.0000075373 | DREF//INSECT BIOMED//DNA REPLICATION RELATED GENE |
| 9 | 0.0000072820 | ANTI THY 12 ANTIBODY//CELL TYPE IDENTITY//ENHANCER CORE |
| 10 | 0.0000072093 | NUSG//TRANSCRIPTION TERMINATION//RHO FACTOR |