Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
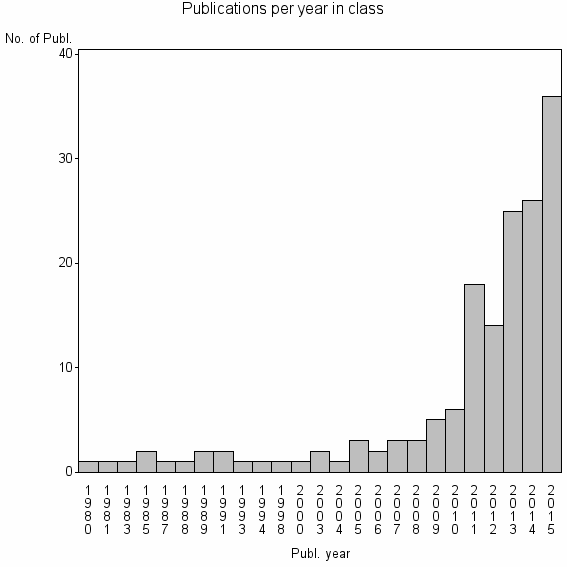
| 29033 | 159 | 38.6 | 59% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 3252 | 1456 | DISSOLVED DNA//NATURAL TRANSFORMATION//EDNA |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | EDNA | Author keyword | 12 | 39% | 15% | 24 |
| 2 | HELLBENDER | Author keyword | 8 | 60% | 6% | 9 |
| 3 | ENVIRONMENTAL DNA | Author keyword | 6 | 23% | 15% | 24 |
| 4 | CRYPTOBRANCHUS | Author keyword | 5 | 60% | 4% | 6 |
| 5 | CRYPTOBRANCHUS ALLEGANIENSIS | Author keyword | 4 | 56% | 3% | 5 |
| 6 | GREAT LAKES PROJECT | Address | 3 | 60% | 2% | 3 |
| 7 | PYTHON MOLURUS BIVITTATUS | Author keyword | 2 | 33% | 3% | 5 |
| 8 | BURMESE PYTHONS | Author keyword | 1 | 100% | 1% | 2 |
| 9 | DNA BASED MONITORING | Author keyword | 1 | 100% | 1% | 2 |
| 10 | BURMESE PYTHON | Author keyword | 1 | 27% | 3% | 4 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | EDNA | 12 | 39% | 15% | 24 | Search EDNA | Search EDNA |
| 2 | HELLBENDER | 8 | 60% | 6% | 9 | Search HELLBENDER | Search HELLBENDER |
| 3 | ENVIRONMENTAL DNA | 6 | 23% | 15% | 24 | Search ENVIRONMENTAL+DNA | Search ENVIRONMENTAL+DNA |
| 4 | CRYPTOBRANCHUS | 5 | 60% | 4% | 6 | Search CRYPTOBRANCHUS | Search CRYPTOBRANCHUS |
| 5 | CRYPTOBRANCHUS ALLEGANIENSIS | 4 | 56% | 3% | 5 | Search CRYPTOBRANCHUS+ALLEGANIENSIS | Search CRYPTOBRANCHUS+ALLEGANIENSIS |
| 6 | PYTHON MOLURUS BIVITTATUS | 2 | 33% | 3% | 5 | Search PYTHON+MOLURUS+BIVITTATUS | Search PYTHON+MOLURUS+BIVITTATUS |
| 7 | BURMESE PYTHONS | 1 | 100% | 1% | 2 | Search BURMESE+PYTHONS | Search BURMESE+PYTHONS |
| 8 | DNA BASED MONITORING | 1 | 100% | 1% | 2 | Search DNA+BASED+MONITORING | Search DNA+BASED+MONITORING |
| 9 | BURMESE PYTHON | 1 | 27% | 3% | 4 | Search BURMESE+PYTHON | Search BURMESE+PYTHON |
| 10 | ENVIRONMENTAL DNA EDNA | 1 | 33% | 1% | 2 | Search ENVIRONMENTAL+DNA+EDNA | Search ENVIRONMENTAL+DNA+EDNA |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | LONG LIVED SALAMANDER | 42 | 94% | 9% | 15 |
| 2 | BISHOPI | 20 | 100% | 6% | 9 |
| 3 | EDNA | 19 | 74% | 9% | 14 |
| 4 | OZARK HELLBENDER | 15 | 77% | 6% | 10 |
| 5 | MOLURUS BIVITTATUS | 2 | 36% | 3% | 5 |
| 6 | CRYPTOBRANCHUS ALLEGANIENSIS BISHOPI | 2 | 67% | 1% | 2 |
| 7 | OZARK | 1 | 38% | 2% | 3 |
| 8 | ANALYTICAL INVERSIONS | 1 | 50% | 1% | 1 |
| 9 | DIRT DNA | 0 | 33% | 1% | 1 |
| 10 | GREAT CRESTED NEWT | 0 | 33% | 1% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| REVIEW The detection of aquatic animal species using environmental DNA - a review of eDNA as a survey tool in ecology | 2014 | 7 | 32 | 75% |
| Environmental DNA for wildlife biology and biodiversity monitoring | 2014 | 26 | 79 | 24% |
| Weighing empirical and hypothetical evidence for assessing potential invasive species range limits: a review of the case of Burmese pythons in the USA | 2014 | 0 | 27 | 37% |
| History, applications, methodological issues and perspectives for the use of environmental DNA (eDNA) in marine and freshwater environments | 2014 | 1 | 47 | 32% |
| The aggressive invasion of exotic reptiles in Florida with a focus on prominent species: A review | 2011 | 5 | 28 | 25% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | GREAT LAKES PROJECT | 3 | 60% | 1.9% | 3 |
| 2 | ADAS UK LTD | 1 | 33% | 1.9% | 3 |
| 3 | ENVIRONM CHANGE INITIAT | 1 | 11% | 6.3% | 10 |
| 4 | AQUAT CONSERVAT | 1 | 20% | 2.5% | 4 |
| 5 | ADAS BOXWORTH | 1 | 33% | 1.3% | 2 |
| 6 | MINNESOTA AQUAT INVAS SPECIES | 1 | 50% | 0.6% | 1 |
| 7 | UMR ECOSYST BIO ERS EVOLUT | 1 | 50% | 0.6% | 1 |
| 8 | WHITNEY GENET | 1 | 50% | 0.6% | 1 |
| 9 | ADAS WOLVERHAMPTON | 1 | 25% | 1.3% | 2 |
| 10 | INVAS SPECIES SCI BRANCH | 0 | 33% | 0.6% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000165212 | DNA BARCODING//BIO ERS ONTARIO//DNA BARCODE |
| 2 | 0.0000143690 | BOIGA IRREGULARIS//BROWN TREESNAKE//BROWN TREE SNAKE |
| 3 | 0.0000128372 | BATRACHOCHYTRIUM DENDROBATIDIS//CHYTRIDIOMYCOSIS//AMPHIBIAN DECLINES |
| 4 | 0.0000126290 | DISSOLVED DNA//NATURAL TRANSFORMATION//DNA PERSISTENCE |
| 5 | 0.0000100676 | SILVER CARP//FILTER FEEDING FISH//LAKE KINNERET |
| 6 | 0.0000092581 | ANCIENT DNA//GEOGENET//SKELETAL PREPARATION |
| 7 | 0.0000089324 | BUFO REGULARIS//LUCKE RENAL CARCINOMA//INTEGRIN ALPHA IIB |
| 8 | 0.0000088198 | NONINVASIVE GENETIC SAMPLING//NON INVASIVE GENETIC SAMPLING//FAECAL DNA |
| 9 | 0.0000086123 | SCHINUS TEREBINTHIFOLIUS//SCHINUS MOLLE//SCHINUS MOLLE L |
| 10 | 0.0000081619 | DIGITAL PCR//DROPLET DIGITAL PCR//SINGLE CELL TRANSCRIPTOMICS |