Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
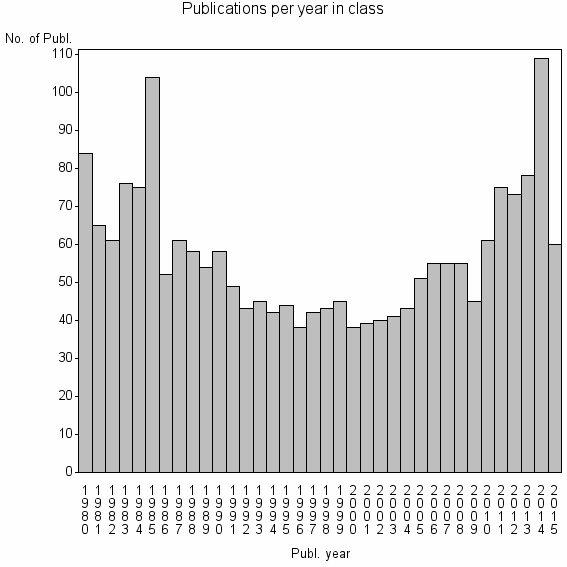
| 2855 | 2057 | 37.4 | 62% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | UMR8041 | Address | 26 | 100% | 1% | 11 |
| 2 | USM0502 | Address | 26 | 87% | 1% | 13 |
| 3 | MUREIN | Author keyword | 22 | 54% | 1% | 28 |
| 4 | PEPTIDOGLYCAN | Author keyword | 19 | 14% | 6% | 120 |
| 5 | PLATEFORME SPE OMETRIE MASSE PROTE MUSEUM | Address | 17 | 100% | 0% | 8 |
| 6 | MOENOMYCIN | Author keyword | 15 | 71% | 1% | 12 |
| 7 | RODZ | Author keyword | 15 | 88% | 0% | 7 |
| 8 | PENICILLIN BINDING PROTEIN | Author keyword | 12 | 26% | 2% | 40 |
| 9 | MREB | Author keyword | 12 | 41% | 1% | 23 |
| 10 | LYTIC TRANSGLYCOSYLASE | Author keyword | 12 | 59% | 1% | 13 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MUREIN | 22 | 54% | 1% | 28 | Search MUREIN | Search MUREIN |
| 2 | PEPTIDOGLYCAN | 19 | 14% | 6% | 120 | Search PEPTIDOGLYCAN | Search PEPTIDOGLYCAN |
| 3 | MOENOMYCIN | 15 | 71% | 1% | 12 | Search MOENOMYCIN | Search MOENOMYCIN |
| 4 | RODZ | 15 | 88% | 0% | 7 | Search RODZ | Search RODZ |
| 5 | PENICILLIN BINDING PROTEIN | 12 | 26% | 2% | 40 | Search PENICILLIN+BINDING+PROTEIN | Search PENICILLIN+BINDING+PROTEIN |
| 6 | MREB | 12 | 41% | 1% | 23 | Search MREB | Search MREB |
| 7 | LYTIC TRANSGLYCOSYLASE | 12 | 59% | 1% | 13 | Search LYTIC+TRANSGLYCOSYLASE | Search LYTIC+TRANSGLYCOSYLASE |
| 8 | PENICILLIN BINDING PROTEINS | 9 | 24% | 2% | 32 | Search PENICILLIN+BINDING+PROTEINS | Search PENICILLIN+BINDING+PROTEINS |
| 9 | BACTERIAL SHAPE | 8 | 75% | 0% | 6 | Search BACTERIAL+SHAPE | Search BACTERIAL+SHAPE |
| 10 | DD CARBOXYPEPTIDASE | 7 | 67% | 0% | 6 | Search DD+CARBOXYPEPTIDASE | Search DD+CARBOXYPEPTIDASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MUREIN | 69 | 58% | 4% | 79 |
| 2 | MUREIN SACCULUS | 65 | 77% | 2% | 44 |
| 3 | ROD SHAPE | 53 | 70% | 2% | 44 |
| 4 | PBP1B | 52 | 100% | 1% | 18 |
| 5 | PENICILLIN BINDING PROTEINS | 49 | 20% | 11% | 219 |
| 6 | BINDING PROTEIN 1B | 44 | 100% | 1% | 16 |
| 7 | GONOCOCCAL PEPTIDOGLYCAN | 36 | 83% | 1% | 20 |
| 8 | MONOFUNCTIONAL GLYCOSYLTRANSFERASE | 36 | 83% | 1% | 20 |
| 9 | BACTERIAL TRANSGLYCOSYLASE | 35 | 89% | 1% | 16 |
| 10 | STRESS BEARING | 30 | 100% | 1% | 12 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| From the regulation of peptidoglycan synthesis to bacterial growth and morphology | 2012 | 156 | 134 | 64% |
| Bacterial morphogenesis and the enigmatic MreB helix | 2015 | 2 | 75 | 81% |
| Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli | 1998 | 561 | 122 | 79% |
| Peptidoglycan structure and architecture | 2008 | 351 | 107 | 65% |
| Different walls for rods and balls: the diversity of peptidoglycan | 2014 | 11 | 75 | 71% |
| Architecture of peptidoglycan: more data and more models | 2010 | 72 | 64 | 83% |
| The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis | 2008 | 255 | 153 | 47% |
| FROM GROWTH TO AUTOLYSIS - THE MUREIN HYDROLASES IN ESCHERICHIA-COLI | 1995 | 91 | 63 | 84% |
| The architecture of the murein (peptidoglycan) in gram-negative bacteria: Vertical scaffold or horizontal layer(s)? | 2004 | 97 | 49 | 86% |
| Structural Perspective of Peptidoglycan Biosynthesis and Assembly | 2012 | 55 | 168 | 46% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | UMR8041 | 26 | 100% | 0.5% | 11 |
| 2 | USM0502 | 26 | 87% | 0.6% | 13 |
| 3 | PLATEFORME SPE OMETRIE MASSE PROTE MUSEUM | 17 | 100% | 0.4% | 8 |
| 4 | BACTERIAL CELL BIOL | 8 | 18% | 1.9% | 40 |
| 5 | RECH DEV ERS MOL | 8 | 100% | 0.2% | 5 |
| 6 | RECH DEV ERSITE MOL | 8 | 100% | 0.2% | 5 |
| 7 | EQUIPE 12 | 5 | 32% | 0.6% | 13 |
| 8 | LRMA | 5 | 21% | 1.0% | 20 |
| 9 | PLATEFORME SPE OMETRIE MASSE PROTEOM MUSEUM | 3 | 100% | 0.1% | 3 |
| 10 | BIOCHIM BACTERIENNE UR477 | 2 | 67% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000194597 | FTSZ//CHLOROPLAST DIVISION//PLASTID DIVISION |
| 2 | 0.0000101171 | NAGZ//AMPG//AMPR |
| 3 | 0.0000101000 | ALANINE RACEMASE//MURD//GLUTAMATE RACEMASE |
| 4 | 0.0000086848 | BETA LACTAMASE//INGN PROT//DD PEPTIDASE |
| 5 | 0.0000076033 | PARB//DNA SEGREGATION//FTSK |
| 6 | 0.0000076026 | CAS 58001 44 8//HOSPITAL PATHOGENS//RV3676 |
| 7 | 0.0000075369 | LIPOTEICHOIC ACID//TEICHOIC ACID//TEICHOIC ACIDS |
| 8 | 0.0000073939 | MECA GENE//MECA//METHICILLIN RESISTANCE |
| 9 | 0.0000070586 | ENDOLYSIN//ENZYBIOTIC//HOLIN |
| 10 | 0.0000067047 | CAULOBACTER//CTRA//CAULOBACTER CRESCENTUS |