Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
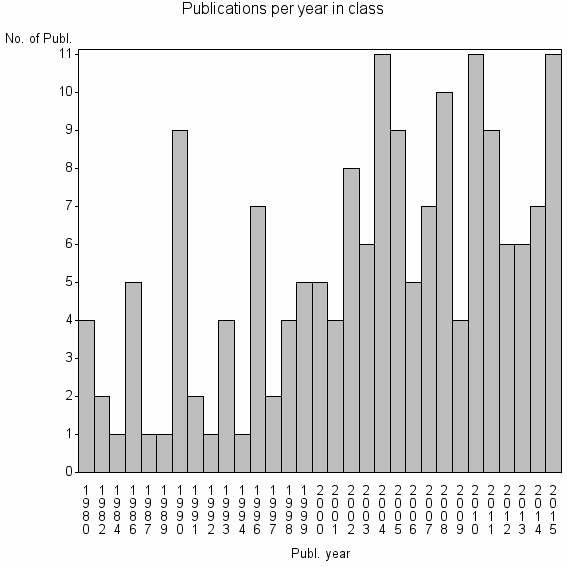
| 28461 | 168 | 37.8 | 81% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | TRNA NUCLEOTIDYLTRANSFERASE | Author keyword | 19 | 80% | 7% | 12 |
| 2 | CCA ADDING ENZYME | Author keyword | 17 | 79% | 7% | 11 |
| 3 | TRNAHIS GUANYLYLTRANSFERASE | Author keyword | 8 | 100% | 3% | 5 |
| 4 | TRNA REPAIR | Author keyword | 6 | 80% | 2% | 4 |
| 5 | CCA ADDITION | Author keyword | 4 | 67% | 2% | 4 |
| 6 | THG1 | Author keyword | 4 | 75% | 2% | 3 |
| 7 | G 1 ADDITION | Author keyword | 3 | 100% | 2% | 3 |
| 8 | TRNA EDITING | Author keyword | 3 | 42% | 3% | 5 |
| 9 | TEMPLATE INDEPENDENT POLYMERIZATION | Author keyword | 2 | 67% | 1% | 2 |
| 10 | NUCLEOTIDYLTRANSFERASE | Author keyword | 2 | 27% | 4% | 7 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TRNA NUCLEOTIDYLTRANSFERASE | 19 | 80% | 7% | 12 | Search TRNA+NUCLEOTIDYLTRANSFERASE | Search TRNA+NUCLEOTIDYLTRANSFERASE |
| 2 | CCA ADDING ENZYME | 17 | 79% | 7% | 11 | Search CCA+ADDING+ENZYME | Search CCA+ADDING+ENZYME |
| 3 | TRNAHIS GUANYLYLTRANSFERASE | 8 | 100% | 3% | 5 | Search TRNAHIS+GUANYLYLTRANSFERASE | Search TRNAHIS+GUANYLYLTRANSFERASE |
| 4 | TRNA REPAIR | 6 | 80% | 2% | 4 | Search TRNA+REPAIR | Search TRNA+REPAIR |
| 5 | CCA ADDITION | 4 | 67% | 2% | 4 | Search CCA+ADDITION | Search CCA+ADDITION |
| 6 | THG1 | 4 | 75% | 2% | 3 | Search THG1 | Search THG1 |
| 7 | G 1 ADDITION | 3 | 100% | 2% | 3 | Search G+1+ADDITION | Search G+1+ADDITION |
| 8 | TRNA EDITING | 3 | 42% | 3% | 5 | Search TRNA+EDITING | Search TRNA+EDITING |
| 9 | TEMPLATE INDEPENDENT POLYMERIZATION | 2 | 67% | 1% | 2 | Search TEMPLATE+INDEPENDENT+POLYMERIZATION | Search TEMPLATE+INDEPENDENT+POLYMERIZATION |
| 10 | NUCLEOTIDYLTRANSFERASE | 2 | 27% | 4% | 7 | Search NUCLEOTIDYLTRANSFERASE | Search NUCLEOTIDYLTRANSFERASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | TRANSFER RNA NUCLEOTIDYLTRANSFERASE | 52 | 74% | 23% | 39 |
| 2 | SACCHAROMYCES CEREVISIAE TRNAHIS | 32 | 85% | 10% | 17 |
| 3 | G 1 ADDITION | 18 | 83% | 6% | 10 |
| 4 | HISTIDINE TRANSFER RNA | 18 | 59% | 12% | 20 |
| 5 | SPIZELLOMYCES PUNCTATUS | 18 | 89% | 5% | 8 |
| 6 | BACTERIAL POLYA POLYMERASE | 17 | 100% | 5% | 8 |
| 7 | CCA ADDING ENZYME | 12 | 40% | 14% | 23 |
| 8 | 5 TERMINUS | 8 | 32% | 12% | 20 |
| 9 | MAMMALIAN POLYA POLYMERASE | 5 | 42% | 6% | 10 |
| 10 | ACANTHAMOEBA CASTELLANII MITOCHONDRIA | 5 | 32% | 8% | 14 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| A story with a good ending: tRNA 3 '-end maturation by CCA-adding enzymes | 2006 | 30 | 16 | 81% |
| Doing it in reverse: 3 '-to-5 ' polymerization by the Thg1 superfamily | 2012 | 12 | 58 | 57% |
| tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization | 2010 | 13 | 78 | 51% |
| tRNA-nucleotidyltransferases: Highly unusual RNA polymerases with vital functions | 2010 | 8 | 42 | 64% |
| RNA-specific ribonucleotidyl transferases | 2007 | 83 | 113 | 14% |
| From End to End: tRNA Editing at 5 '- and 3 '-Terminal Positions | 2014 | 1 | 88 | 42% |
| Controlling translation via modulation of tRNA levels | 2015 | 1 | 134 | 17% |
| This is the end: Processing, editing and repair at the tRNA 3 '-terminus | 2001 | 54 | 58 | 26% |
| Determinants of substrate specificity in RNA-dependent nucleotidyl transferases | 2008 | 9 | 63 | 21% |
| Determinants of tRNA editing and modification: Avoiding conundrums, affecting function | 2012 | 5 | 55 | 24% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | IMMUNODEFICIENCY WALES | 1 | 50% | 0.6% | 1 |
| 2 | INFECT DIS IMMUNODEFICIENCIES UNIT | 1 | 50% | 0.6% | 1 |
| 3 | SYNCHROTRON RADIAT ORGBUNKYO KU | 1 | 50% | 0.6% | 1 |
| 4 | UCD DIABET COMPLICAT | 0 | 33% | 0.6% | 1 |
| 5 | DIPARTMENTO BIOCHIM BIOL MOL ERNESTO QUAGLIARIE | 0 | 25% | 0.6% | 1 |
| 6 | HOP XAVIER BICHAT | 0 | 20% | 0.6% | 1 |
| 7 | OHIO STATE RNA BIOL | 0 | 20% | 0.6% | 1 |
| 8 | PHYSIOL CARDIOVASC GENOM | 0 | 20% | 0.6% | 1 |
| 9 | CHILD HLTH MED | 0 | 14% | 0.6% | 1 |
| 10 | VW GRP | 0 | 13% | 0.6% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000260044 | TRNA SPLICING//POLYNUCLEOTIDE KINASE//SPLICING ENDONUCLEASE |
| 2 | 0.0000207722 | RNASE P//RIBONUCLEASE P//CARTILAGE HAIR HYPOPLASIA |
| 3 | 0.0000203974 | RNASE E//RNASE G//POLYNUCLEOTIDE PHOSPHORYLASE |
| 4 | 0.0000149269 | MAGNETIC ISOTOPE EFFECT//NN VOROZHTZOV ORGAN CHEM//COMPUTAT MODELING GRP |
| 5 | 0.0000145203 | QUEUINE//TRNA MODIFICATION//RNA MODIFICATION |
| 6 | 0.0000133536 | KINETOPLAST//GUIDE RNA//EDITOSOME |
| 7 | 0.0000119729 | AMINOACYL TRNA SYNTHETASE//AMINOACYL TRANSFER RNA SYNTHETASE//AMINOACYLATION |
| 8 | 0.0000100322 | POLYADENYLATION//ALTERNATIVE POLYADENYLATION//POLYA POLYMERASE |
| 9 | 0.0000097641 | LIN28//LIN28B//LIN28A |
| 10 | 0.0000097483 | DAP3//ICT1//MITOCHONDRIAL RIBOSOMAL PROTEINS |