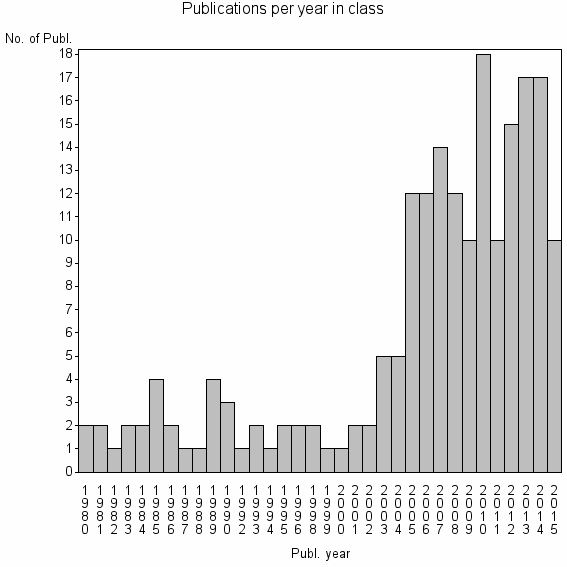
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 27010 | 197 | 46.0 | 70% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 777 | 11593 | MADS BOX GENES//MADS BOX//FLOWER DEVELOPMENT |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PLANT MICROBIAL GENOM | Address | 3 | 60% | 2% | 3 |
| 2 | LRT1 | Author keyword | 2 | 67% | 1% | 2 |
| 3 | NONADDITIVE PROTEIN | Author keyword | 1 | 100% | 1% | 2 |
| 4 | CROP HETEROSIS UTILIZAT MOE | Address | 1 | 19% | 3% | 6 |
| 5 | RUM1 | Author keyword | 1 | 22% | 2% | 4 |
| 6 | BIOINFORMAT BIOMATH GRP | Address | 1 | 50% | 1% | 1 |
| 7 | BIOINFORMAT CHAIR | Address | 1 | 50% | 1% | 1 |
| 8 | CHLOROPHYLL BIOSYNTHETIC PATHWAY | Author keyword | 1 | 50% | 1% | 1 |
| 9 | CROP HETEROSIS | Author keyword | 1 | 50% | 1% | 1 |
| 10 | DIALLELIC CROSSES | Author keyword | 1 | 50% | 1% | 1 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | LRT1 | 2 | 67% | 1% | 2 | Search LRT1 | Search LRT1 |
| 2 | NONADDITIVE PROTEIN | 1 | 100% | 1% | 2 | Search NONADDITIVE+PROTEIN | Search NONADDITIVE+PROTEIN |
| 3 | RUM1 | 1 | 22% | 2% | 4 | Search RUM1 | Search RUM1 |
| 4 | CHLOROPHYLL BIOSYNTHETIC PATHWAY | 1 | 50% | 1% | 1 | Search CHLOROPHYLL+BIOSYNTHETIC+PATHWAY | Search CHLOROPHYLL+BIOSYNTHETIC+PATHWAY |
| 5 | CROP HETEROSIS | 1 | 50% | 1% | 1 | Search CROP+HETEROSIS | Search CROP+HETEROSIS |
| 6 | DIALLELIC CROSSES | 1 | 50% | 1% | 1 | Search DIALLELIC+CROSSES | Search DIALLELIC+CROSSES |
| 7 | EMBRYONIC ROOTS | 1 | 50% | 1% | 1 | Search EMBRYONIC+ROOTS | Search EMBRYONIC+ROOTS |
| 8 | EREBP TRANSCRIPTION FACTOR | 1 | 50% | 1% | 1 | Search EREBP+TRANSCRIPTION+FACTOR | Search EREBP+TRANSCRIPTION+FACTOR |
| 9 | GLOBAL TRANSCRIPT PROFILING | 1 | 50% | 1% | 1 | Search GLOBAL+TRANSCRIPT+PROFILING | Search GLOBAL+TRANSCRIPT+PROFILING |
| 10 | GRASS EMBRYO | 1 | 50% | 1% | 1 | Search GRASS+EMBRYO | Search GRASS+EMBRYO |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | F 1 HYBRID | 19 | 44% | 17% | 34 |
| 2 | PARENTAL INBREDS | 10 | 57% | 6% | 12 |
| 3 | MAYS L F 1 HYBRID | 8 | 100% | 3% | 5 |
| 4 | ELITE RICE HYBRID | 6 | 25% | 11% | 22 |
| 5 | AUTOTETRAPLOID MAIZE | 5 | 55% | 3% | 6 |
| 6 | OVERDOMINANT EPISTATIC LOCI | 5 | 21% | 10% | 19 |
| 7 | WHEAT HYBRID | 4 | 75% | 2% | 3 |
| 8 | INBRED PARENTS | 3 | 100% | 2% | 3 |
| 9 | MUTANT RUM1 | 3 | 100% | 2% | 3 |
| 10 | NONADDITIVE PROTEIN ACCUMULATION | 3 | 100% | 2% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Progress Toward Understanding Heterosis in Crop Plants | 2013 | 38 | 113 | 27% |
| Towards the molecular basis of heterosis | 2007 | 97 | 24 | 54% |
| Genomic and epigenetic insights into the molecular bases of heterosis | 2013 | 41 | 165 | 15% |
| Heterosis: revisiting the magic | 2007 | 165 | 58 | 21% |
| A unifying theory for general multigenic heterosis: energy efficiency, protein metabolism, and implications for molecular breeding | 2011 | 32 | 162 | 14% |
| A Developmental Genetic Basis for Defining Root Classes | 2011 | 4 | 12 | 33% |
| Heterosis in elite hybrid rice: speculation on the genetic and biochemical mechanisms | 2013 | 11 | 50 | 20% |
| Allelic variation and heterosis in maize: How do two halves make more than a whole? | 2007 | 129 | 74 | 15% |
| What is crop heterosis: new insights into an old topic | 2015 | 0 | 93 | 33% |
| Can we learn from heterosis and epigenetics to improve photosynthesis? | 2014 | 0 | 37 | 24% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PLANT MICROBIAL GENOM | 3 | 60% | 1.5% | 3 |
| 2 | CROP HETEROSIS UTILIZAT MOE | 1 | 19% | 3.0% | 6 |
| 3 | BIOINFORMAT BIOMATH GRP | 1 | 50% | 0.5% | 1 |
| 4 | BIOINFORMAT CHAIR | 1 | 50% | 0.5% | 1 |
| 5 | PLANT GENET GERMPLASM ENHANCEMENT | 1 | 50% | 0.5% | 1 |
| 6 | PROTEOM TUEBINGEN | 1 | 50% | 0.5% | 1 |
| 7 | UTILIZAT HETEROSIS IND RICE | 1 | 50% | 0.5% | 1 |
| 8 | CROP FUNCT GENOM | 1 | 29% | 1.0% | 2 |
| 9 | PROTEOME TUEBINGEN | 0 | 11% | 2.0% | 4 |
| 10 | PROTEOME TUBINGEN | 0 | 10% | 2.0% | 4 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000189997 | MAYDICA//POPPING EXPANSION//HETEROTIC GROUPS |
| 2 | 0.0000183381 | MSAP//RNA DIRECTED DNA METHYLATION//RDDM |
| 3 | 0.0000152694 | POLYPLOIDY//AUTOPOLYPLOIDY//ALLOPOLYPLOIDY |
| 4 | 0.0000137435 | RICE GENOME PROGRAM//CROP DESIGN//GENOME IL |
| 5 | 0.0000132507 | ORYZA RUFIPOGON//WIDE COMPATIBILITY//COMMON WILD RICE |
| 6 | 0.0000124559 | EQTL//GENETICAL GENOMICS//ALLELE SPECIFIC GENE EXPRESSION |
| 7 | 0.0000095619 | ANDEAN MAIZE//EMERGENCE FORCE//HETEROTROPHIC AND AUTOTROPHIC GROWTH |
| 8 | 0.0000091382 | PLANT PROTEOMICS//INPPO//LEAF PROTEOME |
| 9 | 0.0000082332 | INVITRO PROTEIN SYNTHESIS//INVIVO PROTEIN SYNTHESIS//AGROSTEMMA GITHAGO L |
| 10 | 0.0000076107 | DOF TRANSCRIPTION FACTOR//DOF1//DOF PROTEIN |