Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
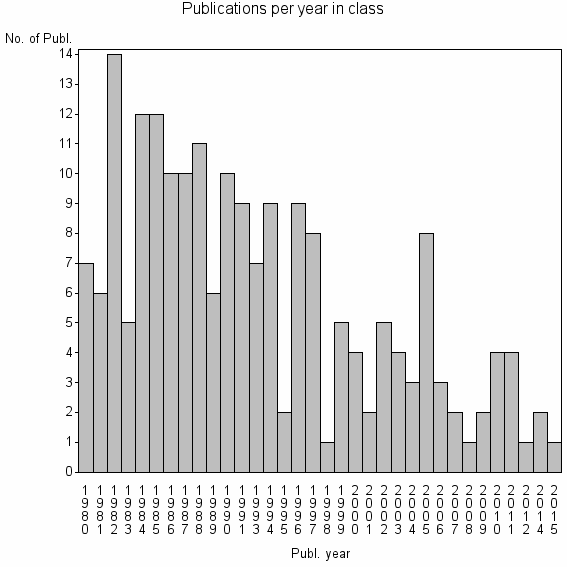
| 26899 | 199 | 25.0 | 33% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1843 | 5621 | RESONANCE RAYLEIGH SCATTERING//RESONANCE LIGHT SCATTERING//DNA BINDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | 5 PHOSPHODIESTERASE | Author keyword | 6 | 71% | 3% | 5 |
| 2 | THREE STRANDED POLYNUCLEOTIDES | Author keyword | 4 | 75% | 2% | 3 |
| 3 | CU ATP COMPLEX | Author keyword | 3 | 100% | 2% | 3 |
| 4 | METAL COMPLEXES OF NUCLEIC ACIDS | Author keyword | 3 | 100% | 2% | 3 |
| 5 | BARLEY MALT SPROUTS | Author keyword | 1 | 100% | 1% | 2 |
| 6 | G C PAIR | Author keyword | 1 | 50% | 1% | 1 |
| 7 | HEMATOPOIETIC STIMULATION | Author keyword | 1 | 50% | 1% | 1 |
| 8 | IONS OF BIVALENT METALS | Author keyword | 1 | 50% | 1% | 1 |
| 9 | 5 RIBONUCLEOTIDES | Author keyword | 1 | 29% | 1% | 2 |
| 10 | NUCLEASE P1 | Author keyword | 1 | 16% | 2% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | 5 PHOSPHODIESTERASE | 6 | 71% | 3% | 5 | Search 5++PHOSPHODIESTERASE | Search 5++PHOSPHODIESTERASE |
| 2 | THREE STRANDED POLYNUCLEOTIDES | 4 | 75% | 2% | 3 | Search THREE+STRANDED+POLYNUCLEOTIDES | Search THREE+STRANDED+POLYNUCLEOTIDES |
| 3 | CU ATP COMPLEX | 3 | 100% | 2% | 3 | Search CU+ATP+COMPLEX | Search CU+ATP+COMPLEX |
| 4 | METAL COMPLEXES OF NUCLEIC ACIDS | 3 | 100% | 2% | 3 | Search METAL+COMPLEXES+OF+NUCLEIC+ACIDS | Search METAL+COMPLEXES+OF+NUCLEIC+ACIDS |
| 5 | BARLEY MALT SPROUTS | 1 | 100% | 1% | 2 | Search BARLEY+MALT+SPROUTS | Search BARLEY+MALT+SPROUTS |
| 6 | G C PAIR | 1 | 50% | 1% | 1 | Search G+C+PAIR | Search G+C+PAIR |
| 7 | HEMATOPOIETIC STIMULATION | 1 | 50% | 1% | 1 | Search HEMATOPOIETIC+STIMULATION | Search HEMATOPOIETIC+STIMULATION |
| 8 | IONS OF BIVALENT METALS | 1 | 50% | 1% | 1 | Search IONS+OF+BIVALENT+METALS | Search IONS+OF+BIVALENT+METALS |
| 9 | 5 RIBONUCLEOTIDES | 1 | 29% | 1% | 2 | Search 5++RIBONUCLEOTIDES | Search 5++RIBONUCLEOTIDES |
| 10 | NUCLEASE P1 | 1 | 16% | 2% | 3 | Search NUCLEASE+P1 | Search NUCLEASE+P1 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | POLY C | 6 | 100% | 2% | 4 |
| 2 | ACID METAL INTERACTIONS | 1 | 100% | 1% | 2 |
| 3 | A POLY U | 1 | 50% | 1% | 1 |
| 4 | POLYRIPOLYRC | 1 | 50% | 1% | 1 |
| 5 | STRANDED POLYRIBONUCLEOTIDES | 1 | 50% | 1% | 1 |
| 6 | BRAIN GLUTAMINASE | 0 | 33% | 1% | 1 |
| 7 | CHRONIC IRRADIATION | 0 | 33% | 1% | 1 |
| 8 | LASER TRANSMITTANCE | 0 | 33% | 1% | 1 |
| 9 | 5 PHOSPHODIESTERASE | 0 | 25% | 1% | 1 |
| 10 | POLYRIBOGUANYLIC ACID | 0 | 25% | 1% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| INTERACTIONS OF METAL-IONS WITH NUCLEIC-ACIDS AND THEIR SUBUNITS - AN ELECTROCHEMICAL APPROACH | 1994 | 29 | 57 | 28% |
| MMR studies of oligonucleotide-metal ion interactions | 1996 | 28 | 39 | 13% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | VERKIN PHYSICOTECH LOW TEMP | 0 | 14% | 1.5% | 3 |
| 2 | BIOMOLEC | 0 | 25% | 0.5% | 1 |
| 3 | VERKIN LOW TEMPERATURE PHYS ENGN | 0 | 20% | 0.5% | 1 |
| 4 | IWERM | 0 | 17% | 0.5% | 1 |
| 5 | HLTH SCI C42 | 0 | 13% | 0.5% | 1 |
| 6 | PHARMACEUT SCI ZHEJIANG | 0 | 100% | 0.5% | 1 |
| 7 | SYDNEY OLYMP PK AUTHOR | 0 | 100% | 0.5% | 1 |
| 8 | U2119 QUAI MONCOUSU | 0 | 100% | 0.5% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000156440 | ANIMAL FEED ENZYMES//CHRYSEOBACTERIUM PROTEOLYTICUM//INTRODUCED PROTEINS |
| 2 | 0.0000150953 | ISOMERIC EQUILIBRIA//CYTIDINE 5 MONOPHOSPHATE//AROMATIC RING STACKING |
| 3 | 0.0000116180 | C 13 LABELED SYNTHONS//BIOENERGY ENVIRONM SCI GRP//DATA PROC PROD GRP |
| 4 | 0.0000113355 | CALF THYMUS DNA//GROOVE BINDING//BINDING SITE SIZE |
| 5 | 0.0000108317 | Z DNA//B Z TRANSITION//Z DNA BINDING PROTEIN |
| 6 | 0.0000107551 | WARWICK ANALYT SCI//GELRED//DNA TOPOISOMERS |
| 7 | 0.0000100449 | KINETICS OF SEPHAROSE STAPHYLOCOCCAL NUCLEASE//SEPHAROSE STAPHYLOCOCCAL NUCLEASE//ELECTROCHEMICAL CELL SENSOR |
| 8 | 0.0000090659 | NUCLEOBASE COMPLEXES//URACIL COMPLEXES//1 METHYLCYTOSINE |
| 9 | 0.0000083506 | DNA CONDENSATION//HIGHER ORDER STRUCTURE OF DNA//LIQUID CRYSTALLINE DISPERSIONS |
| 10 | 0.0000070864 | XANTHINE BIOSENSOR//HYPOXANTHINE//XANTHINE |