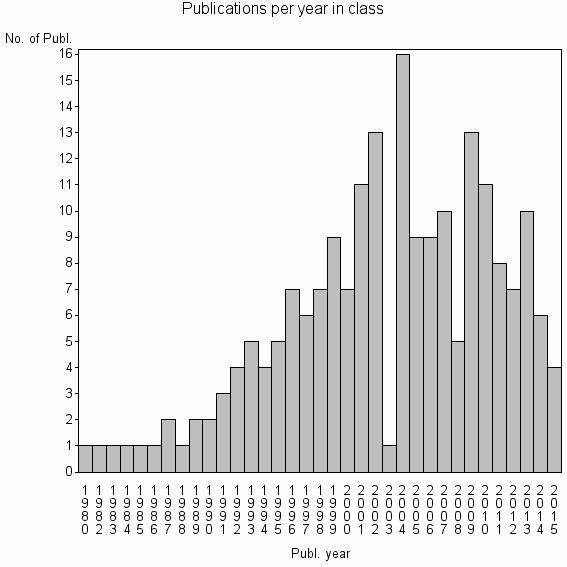
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 26735 | 203 | 35.6 | 69% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1577 | 6675 | NITRILE HYDRATASE//NITRILASE//HYDROGENASE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | 1 5 DIAZACYCLOOCTANE | Author keyword | 3 | 50% | 2% | 5 |
| 2 | 1 4 DIAZACYCLOHEPTANE | Author keyword | 3 | 50% | 2% | 4 |
| 3 | NISOD | Author keyword | 2 | 50% | 1% | 3 |
| 4 | 1 5 DIAZACYCLOOCTANE DACO | Author keyword | 1 | 100% | 1% | 2 |
| 5 | 3D NETWORK ARRAY | Author keyword | 1 | 100% | 1% | 2 |
| 6 | DIAZAMESOCYCLIC LIGAND | Author keyword | 1 | 100% | 1% | 2 |
| 7 | DIAZAMESOCYCLIC LIGANDS | Author keyword | 1 | 100% | 1% | 2 |
| 8 | MESOCYCLIC DIAMINE | Author keyword | 1 | 40% | 1% | 2 |
| 9 | ASSOCIATIVE COMPLEX | Author keyword | 1 | 50% | 0% | 1 |
| 10 | CHAINLIKE STRUCTURE | Author keyword | 1 | 50% | 0% | 1 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | 1 5 DIAZACYCLOOCTANE | 3 | 50% | 2% | 5 | Search 1+5+DIAZACYCLOOCTANE | Search 1+5+DIAZACYCLOOCTANE |
| 2 | 1 4 DIAZACYCLOHEPTANE | 3 | 50% | 2% | 4 | Search 1+4+DIAZACYCLOHEPTANE | Search 1+4+DIAZACYCLOHEPTANE |
| 3 | NISOD | 2 | 50% | 1% | 3 | Search NISOD | Search NISOD |
| 4 | 1 5 DIAZACYCLOOCTANE DACO | 1 | 100% | 1% | 2 | Search 1+5+DIAZACYCLOOCTANE+DACO | Search 1+5+DIAZACYCLOOCTANE+DACO |
| 5 | 3D NETWORK ARRAY | 1 | 100% | 1% | 2 | Search 3D+NETWORK+ARRAY | Search 3D+NETWORK+ARRAY |
| 6 | DIAZAMESOCYCLIC LIGAND | 1 | 100% | 1% | 2 | Search DIAZAMESOCYCLIC+LIGAND | Search DIAZAMESOCYCLIC+LIGAND |
| 7 | DIAZAMESOCYCLIC LIGANDS | 1 | 100% | 1% | 2 | Search DIAZAMESOCYCLIC+LIGANDS | Search DIAZAMESOCYCLIC+LIGANDS |
| 8 | MESOCYCLIC DIAMINE | 1 | 40% | 1% | 2 | Search MESOCYCLIC+DIAMINE | Search MESOCYCLIC+DIAMINE |
| 9 | ASSOCIATIVE COMPLEX | 1 | 50% | 0% | 1 | Search ASSOCIATIVE+COMPLEX | Search ASSOCIATIVE+COMPLEX |
| 10 | CHAINLIKE STRUCTURE | 1 | 50% | 0% | 1 | Search CHAINLIKE+STRUCTURE | Search CHAINLIKE+STRUCTURE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | AMINE AMIDE | 20 | 100% | 4% | 9 |
| 2 | NIN2S2 COMPLEXES | 15 | 77% | 5% | 10 |
| 3 | DIAZAMESOCYCLIC LIGANDS | 8 | 60% | 4% | 9 |
| 4 | 1 5 DIAZACYCLOOCTANE | 6 | 71% | 2% | 5 |
| 5 | HETEROCYCLIC PENDANTS | 6 | 100% | 2% | 4 |
| 6 | NICKEL SUPEROXIDE DISMUTASE | 5 | 50% | 3% | 7 |
| 7 | NISOD | 3 | 100% | 1% | 3 |
| 8 | N N DIMETHYL N N BISBETA MERCAPTOETHYLETHYLENEDIAMINE | 2 | 67% | 1% | 2 |
| 9 | DONOR GROUPS | 2 | 15% | 6% | 13 |
| 10 | NICKEL SITE | 1 | 38% | 1% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Insight into the Structure and Mechanism of Nickel-Containing Superoxide Dismutase Derived from Peptide-Based Mimics | 2014 | 1 | 46 | 59% |
| Oxygen capture by sulfur in nickel thiolates | 1998 | 234 | 48 | 42% |
| COORDINATION CHEMISTRY OF BIDENTATE MEDIUM RING LIGANDS (MESOCYCLES) | 1992 | 61 | 39 | 28% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CRISTALLOCHEM PHYSICOCHIM SOLIDE | 1 | 50% | 0.5% | 1 |
| 2 | RECONNAISSANCE ION CHIM COORDINATSERV CHIM | 1 | 50% | 0.5% | 1 |
| 3 | ZENTRUM MOL BIOMED | 1 | 29% | 1.0% | 2 |
| 4 | CNRS FRE3200 | 0 | 33% | 0.5% | 1 |
| 5 | CRYSTALLOG RMN BIOL | 0 | 33% | 0.5% | 1 |
| 6 | MILENIO MAT COMPLEXOS 2 | 0 | 33% | 0.5% | 1 |
| 7 | PHARM CRISTALLOG RMN BIOL | 0 | 33% | 0.5% | 1 |
| 8 | SERV COMMUN CRISTALLOCHIM | 0 | 25% | 0.5% | 1 |
| 9 | CHIM INORGAN REDOX CHIM MOL GRENOBLE FR | 0 | 20% | 0.5% | 1 |
| 10 | BU S | 0 | 100% | 0.5% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000246066 | SYNGAS FERMENTATION//CLOSTRIDIUM LJUNGDAHLII//ACETYL COA PATHWAY |
| 2 | 0.0000245583 | STEREOSPECIFIC INTERACTIONS//COIII PDII AND COIII PTII COMPLEXES//PENICILLAMINATE COMPLEXES |
| 3 | 0.0000189675 | SUPEROXIDE REDUCTASE//DESULFOFERRODOXIN//RUBRERYTHRIN |
| 4 | 0.0000171536 | NIFE HYDROGENASE//HYDROGENASE//HYDROGENASES |
| 5 | 0.0000160970 | SULFUR LIGAND COMPLEXES//PHOSPHINOTHIOL COMPLEXES//PS LIGANDS |
| 6 | 0.0000156554 | IRON SUPEROXIDE DISMUTASE//SODC//FE SUPEROXIDE DISMUTASE |
| 7 | 0.0000120966 | SOFT SCORPIONATES//AMBIPHILIC LIGANDS//BISMERCAPTOIMIDAZOLYLBORATE COMPLEXES |
| 8 | 0.0000110794 | XRAY PHYS GRP//CIS OCTAHEDRAL GEOMETRY OF NICKELII//N PROPYLETHANE 1 2 DIAMINE |
| 9 | 0.0000097724 | MIXED CHELATE//SELECTIVE ENERGY TRANSFER MODEL//SELECTIVE ENERGY TRANSFER |
| 10 | 0.0000090866 | NITRILE HYDRATASE//NITRILASE//AMIDASE |