Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
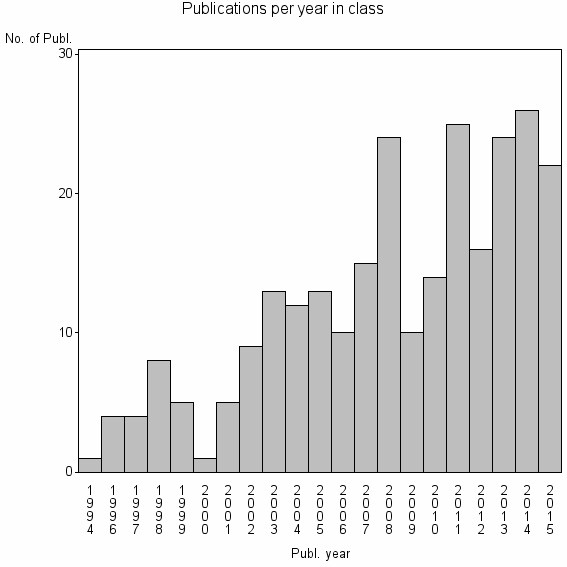
| 24445 | 261 | 46.4 | 94% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | IQGAP1 | Author keyword | 71 | 66% | 25% | 66 |
| 2 | MELUSIN | Author keyword | 24 | 91% | 4% | 10 |
| 3 | IQGAP2 | Author keyword | 20 | 100% | 3% | 9 |
| 4 | IQGAP | Author keyword | 6 | 38% | 5% | 13 |
| 5 | CHP 1 | Author keyword | 6 | 100% | 2% | 4 |
| 6 | IQGAP3 | Author keyword | 3 | 100% | 1% | 3 |
| 7 | IQ DOMAIN GTPASE ACTIVATING PROTEIN 1 | Author keyword | 2 | 67% | 1% | 2 |
| 8 | MORGANA | Author keyword | 2 | 67% | 1% | 2 |
| 9 | IQ MOTIF CONTAINING GTPASE ACTIVATING PROTEIN 1 | Author keyword | 1 | 100% | 1% | 2 |
| 10 | IQG1 | Author keyword | 1 | 50% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | IQGAP1 | 71 | 66% | 25% | 66 | Search IQGAP1 | Search IQGAP1 |
| 2 | MELUSIN | 24 | 91% | 4% | 10 | Search MELUSIN | Search MELUSIN |
| 3 | IQGAP2 | 20 | 100% | 3% | 9 | Search IQGAP2 | Search IQGAP2 |
| 4 | IQGAP | 6 | 38% | 5% | 13 | Search IQGAP | Search IQGAP |
| 5 | CHP 1 | 6 | 100% | 2% | 4 | Search CHP+1 | Search CHP+1 |
| 6 | IQGAP3 | 3 | 100% | 1% | 3 | Search IQGAP3 | Search IQGAP3 |
| 7 | IQ DOMAIN GTPASE ACTIVATING PROTEIN 1 | 2 | 67% | 1% | 2 | Search IQ+DOMAIN+GTPASE+ACTIVATING+PROTEIN+1 | Search IQ+DOMAIN+GTPASE+ACTIVATING+PROTEIN+1 |
| 8 | MORGANA | 2 | 67% | 1% | 2 | Search MORGANA | Search MORGANA |
| 9 | IQ MOTIF CONTAINING GTPASE ACTIVATING PROTEIN 1 | 1 | 100% | 1% | 2 | Search IQ+MOTIF+CONTAINING+GTPASE+ACTIVATING+PROTEIN+1 | Search IQ+MOTIF+CONTAINING+GTPASE+ACTIVATING+PROTEIN+1 |
| 10 | IQG1 | 1 | 50% | 1% | 2 | Search IQG1 | Search IQG1 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RASGAP RELATED PROTEIN | 29 | 71% | 9% | 24 |
| 2 | INVASION FRONTS | 26 | 87% | 5% | 13 |
| 3 | INTEGRATES CA2 CALMODULIN | 17 | 100% | 3% | 8 |
| 4 | IQGAP1 | 10 | 31% | 10% | 26 |
| 5 | MELUSIN | 9 | 83% | 2% | 5 |
| 6 | RAC BOUND BETA 1 INTEGRIN | 5 | 63% | 2% | 5 |
| 7 | CALPONIN HOMOLOGY DOMAIN | 2 | 27% | 3% | 7 |
| 8 | ACTIVATED RECEPTOR 3 | 1 | 100% | 1% | 2 |
| 9 | PROTEIN IQGAP1 | 1 | 100% | 1% | 2 |
| 10 | CAPTURE MICROTUBULES | 1 | 33% | 1% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The biology of IQGAP proteins: beyond the cytoskeleton | 2015 | 2 | 209 | 47% |
| IQGAPs choreograph cellular signaling from the membrane to the nucleus | 2015 | 1 | 135 | 61% |
| IQGAP1 and its binding proteins control diverse biological functions | 2012 | 63 | 114 | 50% |
| IQGAPs in cancer: A family of scaffold proteins underlying tumorigenesis | 2009 | 85 | 98 | 57% |
| IQGAP proteins are integral components of cytoskeletal regulation | 2003 | 153 | 40 | 65% |
| IQGAP1 regulation and roles in cancer | 2009 | 52 | 75 | 64% |
| IQGAP1 in cellular signaling: bridging the GAP | 2006 | 110 | 69 | 48% |
| IQGAP1: A Regulator of Intracellular Spacetime Relativity | 2012 | 11 | 112 | 52% |
| IQGAPs as Key Regulators of Actin-cytoskeleton Dynamics | 2015 | 0 | 65 | 74% |
| IQGAP1 in microbial pathogenesis: Targeting the actin cytoskeleton | 2011 | 11 | 82 | 35% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EMI 0104 | 1 | 33% | 0.8% | 2 |
| 2 | CELL PHARMACOLSHOWA KU | 1 | 50% | 0.4% | 1 |
| 3 | DIPARTIMENTO ANGIOCARDIO NEUROL | 1 | 50% | 0.4% | 1 |
| 4 | IGP PROGRAM | 1 | 50% | 0.4% | 1 |
| 5 | IMMUNOGENET LIG | 1 | 50% | 0.4% | 1 |
| 6 | MONTREAL PROTE NETWORK | 1 | 50% | 0.4% | 1 |
| 7 | NEUROBIOCHEM BIOOPT | 1 | 50% | 0.4% | 1 |
| 8 | PULM DISMINIST HLTH CHINATONGJI HOSP | 1 | 50% | 0.4% | 1 |
| 9 | SMART HIGH THROUGHPUT IL | 1 | 50% | 0.4% | 1 |
| 10 | UNITE 873 | 1 | 50% | 0.4% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000106454 | TIAM1//RHOB//RHOGDI2 |
| 2 | 0.0000092584 | MIG 6//ACK1//RALT |
| 3 | 0.0000089046 | KSR1//PEA 15//IST ENDOCRINOL ONCOL SPERIMENTALE GAETANO SALVA |
| 4 | 0.0000066546 | MAZUMDAR SHAW TRANSLAT//2 DIMENSIONAL LIQUID CHROMATOGRAPHY//BIOLOGICAL VARIATION ANALYSIS |
| 5 | 0.0000064406 | DYNACTIN//CLIP 170//EB1 |
| 6 | 0.0000062795 | CADHERIN//N CADHERIN//P120CTN |
| 7 | 0.0000060458 | RLIP76//RALBP1//RAL |
| 8 | 0.0000060409 | ESCHERICHIA COLI K1//IBEA//E COLI K1 |
| 9 | 0.0000059249 | FORMIN//FHOD1//CELL STRUCT SIGNAL INTEGRAT |
| 10 | 0.0000056187 | CYTOKINESIS//CLEAVAGE FURROW//CENTRALSPINDLIN |