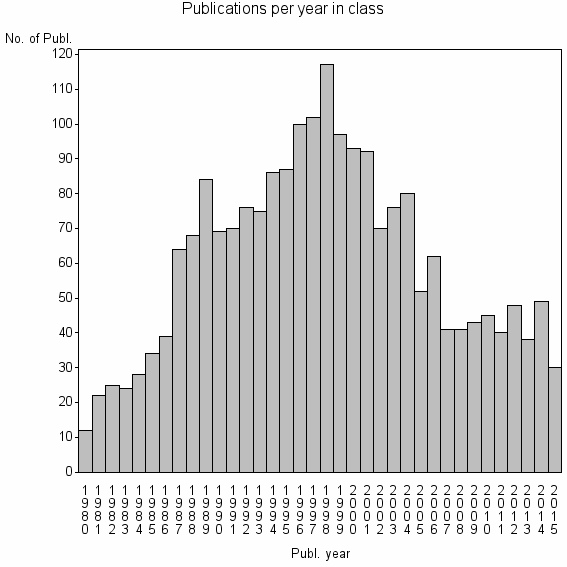
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 2429 | 2179 | 45.5 | 76% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1635 | 6457 | Z DNA//JOURNAL OF BIOMOLECULAR STRUCTURE & DYNAMICS//B Z TRANSITION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | DNA CURVATURE | Author keyword | 25 | 44% | 2% | 43 |
| 2 | A TRACT | Author keyword | 15 | 68% | 1% | 13 |
| 3 | DNA FLEXIBILITY | Author keyword | 14 | 41% | 1% | 26 |
| 4 | SEQUENCE DEPENDENT STRUCTURE | Author keyword | 13 | 80% | 0% | 8 |
| 5 | A TRACTS | Author keyword | 13 | 71% | 0% | 10 |
| 6 | B DNA | Author keyword | 12 | 36% | 1% | 27 |
| 7 | DNA BENDING | Author keyword | 9 | 17% | 2% | 49 |
| 8 | BIOCHIM THEOR | Address | 9 | 17% | 2% | 46 |
| 9 | MOL BIOPHYS PROGRAM | Address | 8 | 20% | 2% | 37 |
| 10 | UPR 9080 | Address | 8 | 21% | 2% | 34 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | DNA CURVATURE | 25 | 44% | 2% | 43 | Search DNA+CURVATURE | Search DNA+CURVATURE |
| 2 | A TRACT | 15 | 68% | 1% | 13 | Search A+TRACT | Search A+TRACT |
| 3 | DNA FLEXIBILITY | 14 | 41% | 1% | 26 | Search DNA+FLEXIBILITY | Search DNA+FLEXIBILITY |
| 4 | SEQUENCE DEPENDENT STRUCTURE | 13 | 80% | 0% | 8 | Search SEQUENCE+DEPENDENT+STRUCTURE | Search SEQUENCE+DEPENDENT+STRUCTURE |
| 5 | A TRACTS | 13 | 71% | 0% | 10 | Search A+TRACTS | Search A+TRACTS |
| 6 | B DNA | 12 | 36% | 1% | 27 | Search B+DNA | Search B+DNA |
| 7 | DNA BENDING | 9 | 17% | 2% | 49 | Search DNA+BENDING | Search DNA+BENDING |
| 8 | MINOR GROOVE WIDTH | 7 | 67% | 0% | 6 | Search MINOR+GROOVE+WIDTH | Search MINOR+GROOVE+WIDTH |
| 9 | BASE STACKING INTERACTIONS | 6 | 71% | 0% | 5 | Search BASE+STACKING+INTERACTIONS | Search BASE+STACKING+INTERACTIONS |
| 10 | DNA HYDRATION | 6 | 42% | 1% | 11 | Search DNA+HYDRATION | Search DNA+HYDRATION |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | B DNA | 196 | 28% | 28% | 607 |
| 2 | DODECAMER | 119 | 52% | 7% | 160 |
| 3 | T G G | 111 | 80% | 3% | 70 |
| 4 | A DNA | 88 | 45% | 7% | 145 |
| 5 | ADENINE TRACT | 67 | 68% | 3% | 58 |
| 6 | CURVED DNA | 66 | 33% | 8% | 165 |
| 7 | C G | 57 | 71% | 2% | 46 |
| 8 | UNIQUE TETRANUCLEOTIDE SEQUENCES | 54 | 68% | 2% | 48 |
| 9 | SEQUENCE DIRECTED CURVATURE | 54 | 80% | 2% | 33 |
| 10 | HELIX GEOMETRY | 41 | 67% | 2% | 37 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| All- Atom Polarizable Force Field for DNA Based on the Classical Drude Oscillator Model | 2014 | 17 | 89 | 45% |
| INTRINSICALLY BENT DNA | 1990 | 376 | 45 | 91% |
| Simulation and modeling of nucleic acid structure, dynamics and interactions | 2004 | 157 | 83 | 58% |
| SEQUENCE-DEPENDENT DNA-STRUCTURE - THE ROLE OF BASE STACKING INTERACTIONS | 1993 | 350 | 90 | 76% |
| Twenty-Five Years of Nucleic Acid Simulations | 2013 | 22 | 72 | 46% |
| DNA curvature and flexibility in vitro and in vivo | 2010 | 66 | 210 | 39% |
| SEQUENCE-DIRECTED CURVATURE OF DNA | 1990 | 407 | 113 | 65% |
| Frontiers in Molecular Dynamics Simulations of DNA | 2012 | 58 | 90 | 34% |
| Molecular dynamics simulation of nucleic acids: Successes, limitations, and promise | 2001 | 165 | 119 | 49% |
| A 5-nanosecond molecular dynamics trajectory for B-DNA: Analysis of structure, motions, and solvation | 1997 | 231 | 93 | 57% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOCHIM THEOR | 9 | 17% | 2.1% | 46 |
| 2 | MOL BIOPHYS PROGRAM | 8 | 20% | 1.7% | 37 |
| 3 | UPR 9080 | 8 | 21% | 1.6% | 34 |
| 4 | BIOL MACROMOL STRUCT | 6 | 44% | 0.5% | 11 |
| 5 | CNRS UPR 9080 | 4 | 44% | 0.3% | 7 |
| 6 | BECKMAN CHEM | 2 | 67% | 0.1% | 2 |
| 7 | JOHNSTON 200 | 2 | 40% | 0.2% | 4 |
| 8 | INFORMAT CHEM | 1 | 25% | 0.2% | 5 |
| 9 | RGFCP | 1 | 50% | 0.1% | 2 |
| 10 | SWISS FED POLYTECH | 1 | 100% | 0.1% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000179011 | Z DNA//B Z TRANSITION//Z DNA BINDING PROTEIN |
| 2 | 0.0000178723 | SHEARED G CENTER DOT A PAIR//DGGTATACC2//SHEARED A CENTER DOT C PAIR |
| 3 | 0.0000136053 | NUCLEOSOME POSITIONING//NUCLEOSOME//NUCLEOSOME MAPPING |
| 4 | 0.0000108230 | TETRAHYDROCORTISOL//OUABAIN BINDING SITE//VEKTOR STATE VIROL BIOTECHNOL |
| 5 | 0.0000107208 | PROGRAM BIOCHEM CHEM//RNA CRYSTALLOGRAPHY//5 METHYL CYTIDINE |
| 6 | 0.0000098205 | REVERSING PULSE ELECTRIC BIREFRINGENCE//LOW FREQUENCY ANOMALIES IN COLLOIDAL ELECTRO OPTICS//ELECTRIC DICHROISM |
| 7 | 0.0000093862 | BMSE PROGRAM//MAGNETIC TWEEZERS//SMALL BIOSYST |
| 8 | 0.0000089052 | BASE PAIR KINETICS//GNRA TETRALOOPS//RAAA HAIRPIN MOTIF |
| 9 | 0.0000088148 | DNA DYNAMICS//PEYRARD BISHOP MODEL//DNA DENATURATION |
| 10 | 0.0000082847 | PROTEIN HYDRATION//INTERNAL SOLVENT//SOLVENT MODELING |