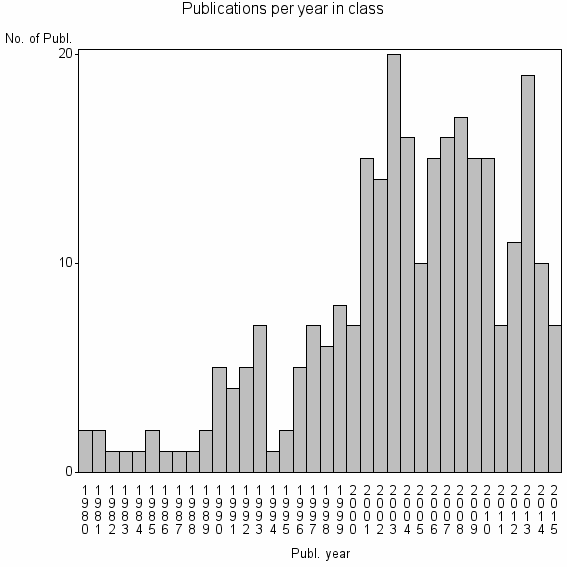
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 23870 | 278 | 38.1 | 74% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | BASE PAIR KINETICS | Author keyword | 1 | 100% | 1% | 2 |
| 2 | GNRA TETRALOOPS | Author keyword | 1 | 50% | 1% | 2 |
| 3 | RAAA HAIRPIN MOTIF | Author keyword | 1 | 100% | 1% | 2 |
| 4 | GNRA TETRALOOP | Author keyword | 1 | 21% | 1% | 3 |
| 5 | BIOENGN M C 063 | Address | 1 | 50% | 0% | 1 |
| 6 | DNA MOLECULAR DYNAMICS | Author keyword | 1 | 50% | 0% | 1 |
| 7 | DSMIPHTURA2306 | Address | 1 | 50% | 0% | 1 |
| 8 | G QUENCHING | Author keyword | 1 | 50% | 0% | 1 |
| 9 | HAIRPIN LOOP DNA | Author keyword | 1 | 50% | 0% | 1 |
| 10 | MACROMOLECULAR FOLDING | Author keyword | 1 | 50% | 0% | 1 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | BASE PAIR KINETICS | 1 | 100% | 1% | 2 | Search BASE+PAIR+KINETICS | Search BASE+PAIR+KINETICS |
| 2 | GNRA TETRALOOPS | 1 | 50% | 1% | 2 | Search GNRA+TETRALOOPS | Search GNRA+TETRALOOPS |
| 3 | RAAA HAIRPIN MOTIF | 1 | 100% | 1% | 2 | Search RAAA+HAIRPIN+MOTIF | Search RAAA+HAIRPIN+MOTIF |
| 4 | GNRA TETRALOOP | 1 | 21% | 1% | 3 | Search GNRA+TETRALOOP | Search GNRA+TETRALOOP |
| 5 | DNA MOLECULAR DYNAMICS | 1 | 50% | 0% | 1 | Search DNA+MOLECULAR+DYNAMICS | Search DNA+MOLECULAR+DYNAMICS |
| 6 | G QUENCHING | 1 | 50% | 0% | 1 | Search G+QUENCHING | Search G+QUENCHING |
| 7 | HAIRPIN LOOP DNA | 1 | 50% | 0% | 1 | Search HAIRPIN+LOOP+DNA | Search HAIRPIN+LOOP+DNA |
| 8 | MACROMOLECULAR FOLDING | 1 | 50% | 0% | 1 | Search MACROMOLECULAR+FOLDING | Search MACROMOLECULAR+FOLDING |
| 9 | NUCLEIC ACID ENERGETICS | 1 | 50% | 0% | 1 | Search NUCLEIC+ACID+ENERGETICS | Search NUCLEIC+ACID+ENERGETICS |
| 10 | UUCG TETRALOOP | 1 | 50% | 0% | 1 | Search UUCG+TETRALOOP | Search UUCG+TETRALOOP |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CONTROLLED INTRACHAIN REACTIONS | 23 | 44% | 14% | 40 |
| 2 | DEPENDENT SURVIVAL PROBABILITY | 14 | 100% | 3% | 7 |
| 3 | POLYMER CYCLIZATION | 11 | 78% | 3% | 7 |
| 4 | INTRAMOLECULAR REACTION RATES | 8 | 75% | 2% | 6 |
| 5 | RUGGED ENERGY LANDSCAPE | 8 | 75% | 2% | 6 |
| 6 | TETRALOOPS | 8 | 50% | 4% | 11 |
| 7 | UNUSUALLY STABLE RNA | 7 | 30% | 8% | 21 |
| 8 | DNA HAIRPIN | 7 | 28% | 8% | 21 |
| 9 | LOOP FORMATION | 6 | 20% | 9% | 25 |
| 10 | INTRAMOLECULAR CONTACT FORMATION | 4 | 19% | 7% | 19 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Kinetics and Dynamics of DNA Hybridization | 2011 | 20 | 37 | 43% |
| LASER-INDUCED TEMPERATURE JUMP INFRARED MEASUREMENTS OF RNA FOLDING | 2009 | 5 | 17 | 65% |
| Loop dependence of the stability and dynamics of nucleic acid hairpins | 2008 | 32 | 106 | 29% |
| Structures, kinetics, thermodynamics, and biological functions of RNA hairpins | 2008 | 37 | 129 | 16% |
| RNA in motion | 2008 | 34 | 27 | 7% |
| Use of RNA-bound Tb3+ as a FRET donor | 2010 | 4 | 32 | 9% |
| EXCEPTIONALLY STABLE NUCLEIC-ACID HAIRPINS | 1995 | 184 | 69 | 9% |
| Internally mismatched RNA: pH and solvent dependence of the thermal unfolding of tRNA(Ala) acceptor stem microhairpins | 2002 | 6 | 66 | 12% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOENGN M C 063 | 1 | 50% | 0.4% | 1 |
| 2 | DSMIPHTURA2306 | 1 | 50% | 0.4% | 1 |
| 3 | GRP PL MATH COMPUTAT BIOL | 0 | 20% | 0.7% | 2 |
| 4 | UP 22 | 0 | 33% | 0.4% | 1 |
| 5 | UMR CNRS 7033 | 0 | 14% | 0.7% | 2 |
| 6 | BIOL CHEM CHEM MOL ENGN | 0 | 25% | 0.4% | 1 |
| 7 | URA 487 | 0 | 13% | 0.7% | 2 |
| 8 | GRP COMPUTAT BIOL PL MATH | 0 | 12% | 0.7% | 2 |
| 9 | BEIJING MOL SCI CHEM BIOL | 0 | 20% | 0.4% | 1 |
| 10 | STATE STRUCT CHEM UNSTABLE STABLE S | 0 | 20% | 0.4% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000174545 | RNA SECONDARY STRUCTURE//RNA SECONDARY STRUCTURE PREDICTION//RNA STRUCTURE PREDICTION |
| 2 | 0.0000166401 | SHEARED G CENTER DOT A PAIR//DGGTATACC2//SHEARED A CENTER DOT C PAIR |
| 3 | 0.0000154641 | BMSE PROGRAM//MAGNETIC TWEEZERS//SMALL BIOSYST |
| 4 | 0.0000132050 | SINGLE MOLECULE ENZYMOLOGY//SINGLE MOLECULE DETECTION//SINGLE MOLECULE STUDIES |
| 5 | 0.0000130950 | RIBOSE AND NUCLEOBASE//ALTERNATE SITE SPECIFIC LABELING//DL323 |
| 6 | 0.0000117172 | NARROW ESCAPE//DEN DANS DM2S SERMA LTSD//NANOMED MECH BIOL |
| 7 | 0.0000106415 | PROTEIN FOLDING//PHI VALUE//CONTACT ORDER |
| 8 | 0.0000100337 | GROUP I INTRON//SELF SPLICING//RIBOZYME |
| 9 | 0.0000089052 | DNA CURVATURE//A TRACT//DNA FLEXIBILITY |
| 10 | 0.0000085788 | ELECTROSTATIC RATE ENHANCEMENT//ELECTROSTATIC STEERING//WEIGHTED ENSEMBLE BROWNIAN DYNAMICS |