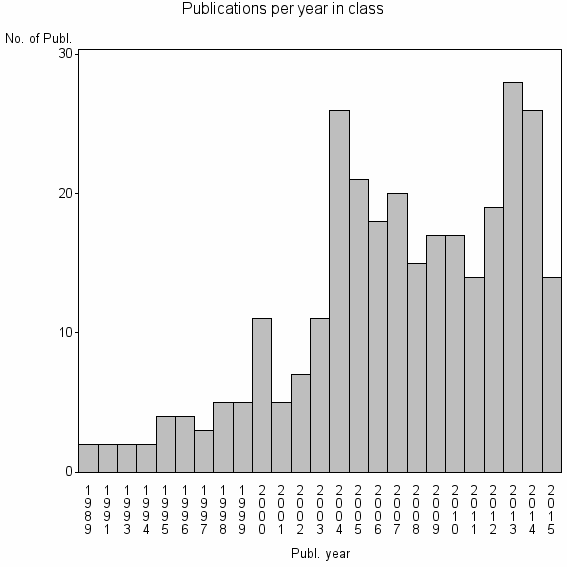
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 23237 | 298 | 41.1 | 86% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 719 | 12132 | SUPRACHIASMATIC NUCLEUS//CIRCADIAN RHYTHMS//CIRCADIAN CLOCK |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | KAIC | Author keyword | 44 | 100% | 5% | 16 |
| 2 | ELECT ENGN BIOL SCI | Address | 17 | 100% | 3% | 8 |
| 3 | KAI GENES | Author keyword | 8 | 100% | 2% | 5 |
| 4 | KAIA | Author keyword | 6 | 80% | 1% | 4 |
| 5 | SYST BIOL MOL CELLULAR BIOL | Address | 4 | 67% | 1% | 4 |
| 6 | QUANTITAT SYST BIOL GRP | Address | 4 | 75% | 1% | 3 |
| 7 | KAIB | Author keyword | 3 | 100% | 1% | 3 |
| 8 | KAIC PHOSPHORYLATION | Author keyword | 3 | 100% | 1% | 3 |
| 9 | KAI | Author keyword | 3 | 33% | 3% | 8 |
| 10 | KAIABC | Author keyword | 3 | 60% | 1% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | KAIC | 44 | 100% | 5% | 16 | Search KAIC | Search KAIC |
| 2 | KAI GENES | 8 | 100% | 2% | 5 | Search KAI+GENES | Search KAI+GENES |
| 3 | KAIA | 6 | 80% | 1% | 4 | Search KAIA | Search KAIA |
| 4 | KAIB | 3 | 100% | 1% | 3 | Search KAIB | Search KAIB |
| 5 | KAIC PHOSPHORYLATION | 3 | 100% | 1% | 3 | Search KAIC+PHOSPHORYLATION | Search KAIC+PHOSPHORYLATION |
| 6 | KAI | 3 | 33% | 3% | 8 | Search KAI | Search KAI |
| 7 | KAIABC | 3 | 60% | 1% | 3 | Search KAIABC | Search KAIABC |
| 8 | LDPA | 2 | 67% | 1% | 2 | Search LDPA | Search LDPA |
| 9 | MONITORING APPARATUS | 2 | 67% | 1% | 2 | Search MONITORING+APPARATUS | Search MONITORING+APPARATUS |
| 10 | CIKA | 1 | 50% | 1% | 2 | Search CIKA | Search CIKA |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | KAIC PHOSPHORYLATION | 46 | 47% | 24% | 71 |
| 2 | PROTEIN KAIA | 29 | 88% | 5% | 14 |
| 3 | SYNECHOCOCCUS ELONGATUS | 24 | 23% | 32% | 94 |
| 4 | SYNECHOCOCCUS ELONGATUS PCC 7942 | 24 | 51% | 11% | 34 |
| 5 | CIKA | 24 | 91% | 3% | 10 |
| 6 | CLOCK SYSTEM | 15 | 82% | 3% | 9 |
| 7 | PSEUDO RECEIVER DOMAIN | 15 | 77% | 3% | 10 |
| 8 | KAIABC | 14 | 100% | 2% | 7 |
| 9 | SASA | 12 | 63% | 4% | 12 |
| 10 | CLOCK PROTEIN KAIA | 11 | 78% | 2% | 7 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| How a cyanobacterium tells time | 2008 | 57 | 53 | 92% |
| Metabolic Compensation and Circadian Resilience in Prokaryotic Cyanobacteria | 2014 | 5 | 136 | 63% |
| The cyanobacterial clock and metabolism | 2014 | 3 | 41 | 88% |
| The Cyanobacterial Circadian System: From Biophysics to Bioevolution | 2011 | 35 | 106 | 85% |
| A cyanobacterial circadian clockwork | 2008 | 34 | 72 | 82% |
| Structural Insights into a Circadian Oscillator | 2008 | 24 | 39 | 92% |
| Winding up the cyanobacterial circandian clock | 2007 | 26 | 48 | 92% |
| Diversity of KaiC-based timing systems in marine Cyanobacteria | 2014 | 2 | 151 | 58% |
| The Itty-Bitty Time Machine: Genetics of the Cyanobacterial Circadian Clock | 2011 | 14 | 124 | 77% |
| Structural and dynamic aspects of protein clocks: how can they be so slow and stable? | 2012 | 5 | 68 | 79% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ELECT ENGN BIOL SCI | 17 | 100% | 2.7% | 8 |
| 2 | SYST BIOL MOL CELLULAR BIOL | 4 | 67% | 1.3% | 4 |
| 3 | QUANTITAT SYST BIOL GRP | 4 | 75% | 1.0% | 3 |
| 4 | CIRCADIAN BIOL | 2 | 29% | 1.7% | 5 |
| 5 | ADV ANAL TECHNOL DEV | 1 | 100% | 0.7% | 2 |
| 6 | INTRACELLULAR REGULAT | 1 | 50% | 0.7% | 2 |
| 7 | MOL POPULAT GENET EVOLUT | 1 | 100% | 0.7% | 2 |
| 8 | SYST BIOL CHEM CHEM BIOL | 1 | 100% | 0.7% | 2 |
| 9 | BIOORIENTED TECHNOL ADVANCEMENT | 1 | 25% | 1.3% | 4 |
| 10 | BIOL CLOCKS | 1 | 11% | 3.0% | 9 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000250863 | PSEUDO RESPONSE REGULATOR//CIRCADIAN CLOCK//GIGANTEA |
| 2 | 0.0000203430 | FRQ//WC 1//WHITE COLLAR 1 |
| 3 | 0.0000198638 | TRICHODESMIUM//CROCOSPHAERA//TRICHODESMIUM SPP |
| 4 | 0.0000177175 | NTCA//BIOQUIM VEGETAL FOTOSINTESIS//HETEROCYST |
| 5 | 0.0000114045 | PIGMENT DISPERSING FACTOR//TIMELESS//ACCESSORY MEDULLA |
| 6 | 0.0000103778 | CLOCK GENES//CLOCK GENE//BMAL1 |
| 7 | 0.0000074426 | EVOLUTION CANYON//NALASSUS//BIODIVERSITY CHARACTERISTICS |
| 8 | 0.0000066495 | PROCHLOROCOCCUS//SYNECHOCOCCUS//PICOPLANKTON |
| 9 | 0.0000037293 | LINEAR NOISE APPROXIMATION//STOCHASTIC GENE EXPRESSION//CHEMICAL MASTER EQUATION |
| 10 | 0.0000032413 | ERATO COMPLEX SYST BIOL//ISOLOGOUS DIVERSIFICATION//COHERENT SPIKING |