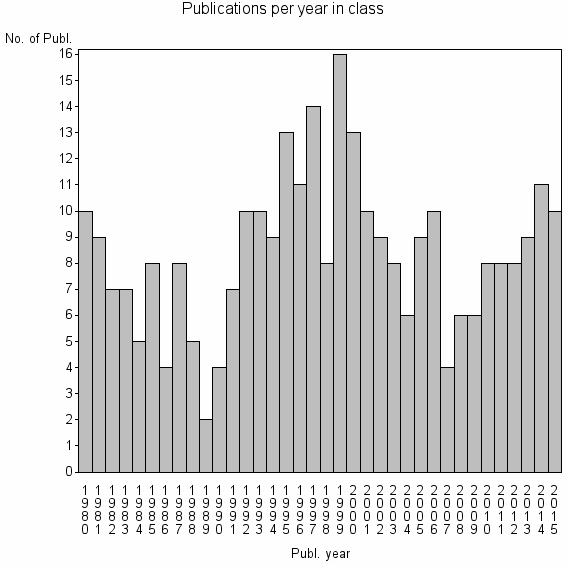
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 23107 | 302 | 36.2 | 70% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | TRANSHYDROGENASE | Author keyword | 90 | 69% | 25% | 77 |
| 2 | NICOTINAMIDE NUCLEOTIDE | Author keyword | 9 | 64% | 3% | 9 |
| 3 | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE | Author keyword | 9 | 67% | 3% | 8 |
| 4 | PYRIDINE NUCLEOTIDE TRANSHYDROGENASE | Author keyword | 7 | 64% | 2% | 7 |
| 5 | PROTON TRANSLOCATION | Author keyword | 3 | 13% | 8% | 24 |
| 6 | RHODOSPIRILLUM RUBRUM | Author keyword | 2 | 11% | 5% | 15 |
| 7 | JR GRP ANIM MODELS OBES 2 | Address | 2 | 43% | 1% | 3 |
| 8 | TRANSHYDROGENATION | Author keyword | 2 | 43% | 1% | 3 |
| 9 | C57BL 6NCRL | Author keyword | 1 | 50% | 1% | 2 |
| 10 | CYCLIC TRANSHYDROGENATION | Author keyword | 1 | 100% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE | 58 | 49% | 29% | 87 |
| 2 | ESCHERICHIA COLI TRANSHYDROGENASE | 53 | 95% | 6% | 18 |
| 3 | TRANSLOCATING TRANSHYDROGENASE | 49 | 94% | 6% | 17 |
| 4 | ATP DRIVEN TRANSHYDROGENASE | 42 | 94% | 5% | 15 |
| 5 | HEART MITOCHONDRIAL TRANSHYDROGENASE | 41 | 100% | 5% | 15 |
| 6 | MOBILE LOOP REGION | 41 | 100% | 5% | 15 |
| 7 | NADPH BINDING COMPONENT DIII | 33 | 100% | 4% | 13 |
| 8 | PROTON TRANSLOCATING TRANSHYDROGENASE | 21 | 69% | 6% | 18 |
| 9 | H TRANSHYDROGENASE | 17 | 100% | 3% | 8 |
| 10 | RECONSTITUTED TRANSHYDROGENASE | 14 | 100% | 2% | 7 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Proton translocation by transhydrogenase | 2003 | 42 | 42 | 81% |
| Nicotinamide nucleotide transhydrogenase: A model for utilization of substrate binding energy for proton translocation | 1996 | 59 | 37 | 92% |
| A review of the binding-change mechanism for proton-translocating transhydrogenase | 2012 | 5 | 78 | 97% |
| Structure and mechanism of proton-translocating transhydrogenase | 1999 | 21 | 59 | 92% |
| Review and Hypothesis. New insights into the reaction mechanism of transhydrogenase: Swivelling the dIII component may gate the proton channel | 2015 | 0 | 54 | 85% |
| THE PROTON-TRANSLOCATING NICOTINAMIDE ADENINE-DINUCLEOTIDE TRANSHYDROGENASE | 1991 | 51 | 46 | 72% |
| MOLECULAR-BIOLOGY OF NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE - A UNIQUE PROTON PUMP | 1995 | 41 | 77 | 53% |
| Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice | 2010 | 79 | 52 | 15% |
| NIH experiment in centralized mouse phenotyping: the Vanderbilt experience and recommendations for evaluating glucose homeostasis in the mouse | 2009 | 34 | 23 | 17% |
| Parallel universes of Black Six biology | 2014 | 1 | 65 | 15% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | JR GRP ANIM MODELS OBES 2 | 2 | 43% | 1.0% | 3 |
| 2 | MOLEC EXPTL MED BIOCHEM | 1 | 14% | 1.7% | 5 |
| 3 | EMILY COUR CLIN CANC | 1 | 50% | 0.3% | 1 |
| 4 | HUMAN GENET UNITIGMM | 1 | 50% | 0.3% | 1 |
| 5 | INTERDISCIPLINARY CARDIOVASC SCI MED | 1 | 50% | 0.3% | 1 |
| 6 | NIH MOUSE METAB PHENOTYPING | 1 | 50% | 0.3% | 1 |
| 7 | VANDERBILT NIDDK MOUSE METAB PHENOTYPING | 1 | 50% | 0.3% | 1 |
| 8 | ARRHENIUS BIOCHEM | 0 | 33% | 0.3% | 1 |
| 9 | MITOCHONDRIAL MOL MED GENET | 0 | 33% | 0.3% | 1 |
| 10 | VANDERBILT NIH MOUSE METAB PHENOTYPING | 0 | 33% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000082700 | PEPEROMIA METALLICA//LIPID RADICAL CYCLES//LIPOPHILIC AMINES |
| 2 | 0.0000077055 | NSY MOUSE//NEW ZEALAND OBESE MOUSE//MOUSE GENOM OURCE |
| 3 | 0.0000076475 | NAD KINASE//GRP BIOCHIM BIOL MOL VEGETALES//NADH KINASE |
| 4 | 0.0000073155 | TRIPLE A SYNDROME//ALACRIMA//ALLGROVE SYNDROME |
| 5 | 0.0000063536 | SPATIAL AND VISUAL DISCRIMINATION//ANIM SCI GENET//CORE UNIT BIOMED |
| 6 | 0.0000060524 | REVERSE ELECTRON FLOW//NEUROBIOCHEM GRP//BRAIN MITOCHONDRION |
| 7 | 0.0000048735 | COMPLEX I//NADH UBIQUINONE OXIDOREDUCTASE//NADH UBIQUINONE OXIDOREDUCTASE |
| 8 | 0.0000045438 | ELECTRON OPTICAL DATA//AXIAL OSTEOMALACIA//FIBROGENESIS IMPERFECTA OSSIUM |
| 9 | 0.0000043429 | 3 ISOPROPYLMALATE DEHYDROGENASE//ISOCITRATE DEHYDROGENASE//ISOPROPYLMALATE DEHYDROGENASE |
| 10 | 0.0000041928 | MONOSODIUM SALT OF METHYLARSONIC ACID//AMIDE PROTON//WEED BIOTYPES |