Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
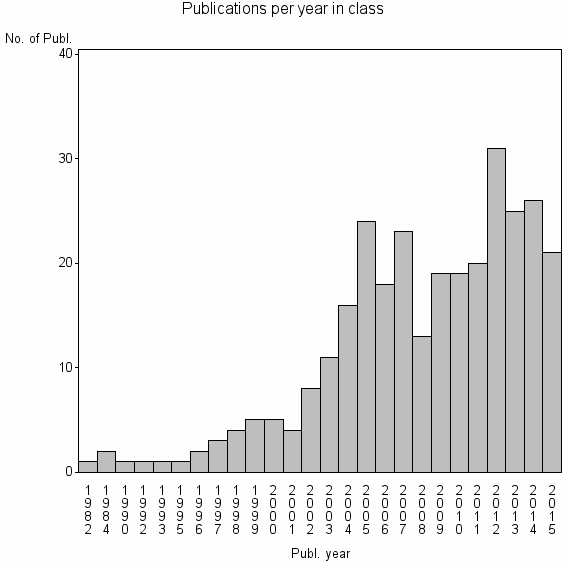
| 23053 | 304 | 28.8 | 47% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 396 | 16220 | CREDIT SCORING//MACHINE LEARNING//FEATURE SELECTION |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MISSING VALUE IMPUTATION | Author keyword | 5 | 37% | 3% | 10 |
| 2 | MISSING VALUE ESTIMATION | Author keyword | 4 | 38% | 3% | 8 |
| 3 | COMPUTAT MATH BIOL | Address | 3 | 40% | 2% | 6 |
| 4 | MISSING DATA IMPUTATION | Author keyword | 3 | 23% | 3% | 10 |
| 5 | AUTOMATIC CLUSTERING ALGORITHM | Author keyword | 2 | 67% | 1% | 2 |
| 6 | MISSING GENES | Author keyword | 2 | 67% | 1% | 2 |
| 7 | CLOSEST FIT | Author keyword | 1 | 50% | 1% | 2 |
| 8 | ELECT ENGN BUILT ENVIRONM | Address | 1 | 50% | 1% | 2 |
| 9 | HOT DECK MODEL | Author keyword | 1 | 100% | 1% | 2 |
| 10 | ITERATIVE IMPUTATION | Author keyword | 1 | 50% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MISSING VALUE IMPUTATION | 5 | 37% | 3% | 10 | Search MISSING+VALUE+IMPUTATION | Search MISSING+VALUE+IMPUTATION |
| 2 | MISSING VALUE ESTIMATION | 4 | 38% | 3% | 8 | Search MISSING+VALUE+ESTIMATION | Search MISSING+VALUE+ESTIMATION |
| 3 | MISSING DATA IMPUTATION | 3 | 23% | 3% | 10 | Search MISSING+DATA+IMPUTATION | Search MISSING+DATA+IMPUTATION |
| 4 | AUTOMATIC CLUSTERING ALGORITHM | 2 | 67% | 1% | 2 | Search AUTOMATIC+CLUSTERING+ALGORITHM | Search AUTOMATIC+CLUSTERING+ALGORITHM |
| 5 | MISSING GENES | 2 | 67% | 1% | 2 | Search MISSING+GENES | Search MISSING+GENES |
| 6 | CLOSEST FIT | 1 | 50% | 1% | 2 | Search CLOSEST+FIT | Search CLOSEST+FIT |
| 7 | HOT DECK MODEL | 1 | 100% | 1% | 2 | Search HOT+DECK+MODEL | Search HOT+DECK+MODEL |
| 8 | ITERATIVE IMPUTATION | 1 | 50% | 1% | 2 | Search ITERATIVE+IMPUTATION | Search ITERATIVE+IMPUTATION |
| 9 | MEAN MODE MODEL | 1 | 100% | 1% | 2 | Search MEAN+MODE+MODEL | Search MEAN+MODE+MODEL |
| 10 | RELATIONAL DATABASE SYSTEMS | 1 | 24% | 2% | 5 | Search RELATIONAL+DATABASE+SYSTEMS | Search RELATIONAL+DATABASE+SYSTEMS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | LEAST SQUARES IMPUTATION | 33 | 100% | 4% | 13 |
| 2 | ESTIMATING NULL VALUES | 4 | 67% | 1% | 4 |
| 3 | MISSING VALUE ESTIMATION | 4 | 26% | 4% | 12 |
| 4 | VALUE IMPUTATION | 3 | 50% | 1% | 4 |
| 5 | WEIGHTED FUZZY RULES | 1 | 100% | 1% | 2 |
| 6 | BIOLOGICAL KNOWLEDGE | 1 | 20% | 2% | 5 |
| 7 | RELATIONAL DATABASE SYSTEMS | 1 | 50% | 0% | 1 |
| 8 | YEAST CULTURES | 1 | 25% | 1% | 2 |
| 9 | BAYESIAN PCA | 0 | 33% | 0% | 1 |
| 10 | BICLUSTERING ALGORITHM | 0 | 33% | 0% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| A Review on Missing Value Imputation Algorithms for Microarray Gene Expression Data | 2014 | 4 | 21 | 71% |
| A Primer on Effectiveness and Efficacy Trials | 2014 | 14 | 24 | 4% |
| Information enhancement for data mining | 2011 | 0 | 12 | 42% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | COMPUTAT MATH BIOL | 3 | 40% | 2.0% | 6 |
| 2 | ELECT ENGN BUILT ENVIRONM | 1 | 50% | 0.7% | 2 |
| 3 | DPTO TECNOL INFORMAC COMUNICAC | 1 | 33% | 0.7% | 2 |
| 4 | CRUSTAL GEOPHYS GEOCHEM SCI | 1 | 12% | 2.0% | 6 |
| 5 | DPTO TEORIA SENAL COMUN | 1 | 50% | 0.3% | 1 |
| 6 | EIF | 1 | 50% | 0.3% | 1 |
| 7 | ELECT INFORMAT ENGN EIE | 1 | 50% | 0.3% | 1 |
| 8 | EPS POLITECN | 1 | 50% | 0.3% | 1 |
| 9 | INFORMAT MANAGMENT ENGN | 1 | 50% | 0.3% | 1 |
| 10 | INSERMUMR S 665 | 1 | 50% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000170641 | BICLUSTERING//GENE EXPRESSION DATA//GENE EXPRESSION DATA ANALYSIS |
| 2 | 0.0000122063 | TAISHO FUNCT GENOM//MULTIPHOTON DETECTION//ADAPTER TAGGED COMPETITIVE PCR |
| 3 | 0.0000117709 | MULTIPLE IMPUTATION//MISSING DATA//PATTERN MIXTURE MODEL |
| 4 | 0.0000094476 | OTONEUROLOGY//VESTIBULAR DIAGNOSIS//OTONEUROLOGICAL DATA |
| 5 | 0.0000074402 | SPORTS PERFORMANCE ANALYSIS//BIOSIGNAL INTERPRETATION//CASEMIX BASED FUNDING |
| 6 | 0.0000070399 | GENE SELECTION//FEATURE SELECTION//CANCER CLASSIFICATION |
| 7 | 0.0000067850 | STUDY VARIABLE//STUDY VARIATE//AUXILIARY VARIABLE |
| 8 | 0.0000066777 | CLASS IMBALANCE PROBLEM//CLASS IMBALANCE//COST SENSITIVE LEARNING |
| 9 | 0.0000063376 | TRANSFER LEARNING//MULTITASK LEARNING//DOMAIN ADAPTATION |
| 10 | 0.0000062017 | PROTOTYPE SELECTION//INSTANCE SELECTION//PROTOTYPE REDUCTION SCHEMES PRS |