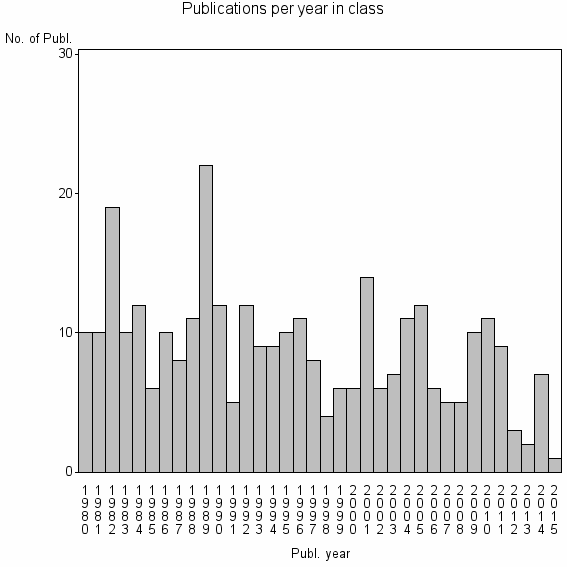
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 22605 | 319 | 29.4 | 61% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MICROSOMAL GLUTATHIONE TRANSFERASE | Author keyword | 53 | 95% | 6% | 18 |
| 2 | MGST1 | Author keyword | 27 | 92% | 3% | 11 |
| 3 | MOL GENET PHARMACOL | Address | 8 | 75% | 2% | 6 |
| 4 | MICROSOMAL GLUTATHIONE S TRANSFERASE | Author keyword | 4 | 46% | 2% | 6 |
| 5 | MICROSOMAL GLUTATHIONE TRANSFERASE 1 | Author keyword | 3 | 57% | 1% | 4 |
| 6 | MGST | Author keyword | 3 | 100% | 1% | 3 |
| 7 | MAPEG | Author keyword | 3 | 42% | 2% | 5 |
| 8 | CU2 ASCORBATE | Author keyword | 2 | 67% | 1% | 2 |
| 9 | FUNCT MOL PHARMACOL | Address | 2 | 67% | 1% | 2 |
| 10 | MGST2 | Author keyword | 2 | 67% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | MICROSOMAL GLUTATHIONE TRANSFERASE | 53 | 95% | 6% | 18 | Search MICROSOMAL+GLUTATHIONE+TRANSFERASE | Search MICROSOMAL+GLUTATHIONE+TRANSFERASE |
| 2 | MGST1 | 27 | 92% | 3% | 11 | Search MGST1 | Search MGST1 |
| 3 | MICROSOMAL GLUTATHIONE S TRANSFERASE | 4 | 46% | 2% | 6 | Search MICROSOMAL+GLUTATHIONE+S+TRANSFERASE | Search MICROSOMAL+GLUTATHIONE+S+TRANSFERASE |
| 4 | MICROSOMAL GLUTATHIONE TRANSFERASE 1 | 3 | 57% | 1% | 4 | Search MICROSOMAL+GLUTATHIONE+TRANSFERASE+1 | Search MICROSOMAL+GLUTATHIONE+TRANSFERASE+1 |
| 5 | MGST | 3 | 100% | 1% | 3 | Search MGST | Search MGST |
| 6 | MAPEG | 3 | 42% | 2% | 5 | Search MAPEG | Search MAPEG |
| 7 | CU2 ASCORBATE | 2 | 67% | 1% | 2 | Search CU2+ASCORBATE | Search CU2+ASCORBATE |
| 8 | MGST2 | 2 | 67% | 1% | 2 | Search MGST2 | Search MGST2 |
| 9 | MGST3 | 2 | 67% | 1% | 2 | Search MGST3 | Search MGST3 |
| 10 | MICROSOMAL GLUTATHIONE S TRANSFERASE 1 | 2 | 67% | 1% | 2 | Search MICROSOMAL+GLUTATHIONE+S+TRANSFERASE+1 | Search MICROSOMAL+GLUTATHIONE+S+TRANSFERASE+1 |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MAPEG | 11 | 78% | 2% | 7 |
| 2 | 6 ANGSTROM RESOLUTION | 9 | 67% | 3% | 8 |
| 3 | DEPENDENT PROTECTION | 5 | 24% | 5% | 17 |
| 4 | MICROSOMAL GLUTATHIONE | 3 | 100% | 1% | 3 |
| 5 | MEMBRANE ASSOCIATED PROTEINS | 3 | 23% | 4% | 12 |
| 6 | MICROSOMAL UDP GLUCURONOSYLTRANSFERASE | 2 | 43% | 1% | 3 |
| 7 | RICH SRCR DOMAIN | 1 | 50% | 1% | 2 |
| 8 | MICROSOMAL GLUTATHIONE TRANSFERASE | 1 | 19% | 2% | 6 |
| 9 | LABILE FACTOR | 1 | 40% | 1% | 2 |
| 10 | CONJUGATE EFFLUX | 1 | 50% | 0% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Microsomal glutathione transferase 1: mechanism and functional roles | 2011 | 17 | 76 | 49% |
| Reactive intermediates and the dynamics of glutathione transferases | 2002 | 78 | 58 | 40% |
| Microsomal glutathione transferase 1 | 2005 | 11 | 27 | 89% |
| Catalysis within the lipid bilayer - structure and mechanism of the MAPEG family of integral membrane proteins | 2008 | 18 | 19 | 47% |
| Mitochondrial glutathione transferases involving a new function for membrane permeability transition pore regulation | 2011 | 12 | 86 | 29% |
| The structure of membrane associated proteins in eicosanoid and glutathione metabolism as determined by electron crystallography | 2007 | 11 | 49 | 22% |
| Two-dimensional crystallization and electron crystallography of MAPEG proteins | 2005 | 3 | 18 | 28% |
| Japanese single nucleotide polymorphism database for 267 possible drug-related genes | 2006 | 8 | 36 | 19% |
| Enzymes without borders: Mobilizing substrates, delivering products | 2008 | 35 | 49 | 6% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MOL GENET PHARMACOL | 8 | 75% | 1.9% | 6 |
| 2 | FUNCT MOL PHARMACOL | 2 | 67% | 0.6% | 2 |
| 3 | RIKEN SNP | 1 | 38% | 0.9% | 3 |
| 4 | DMPK DISCOVERY RD | 1 | 100% | 0.6% | 2 |
| 5 | GRP PERSONALIZED MED SNP ANAL | 1 | 100% | 0.6% | 2 |
| 6 | PHARMACOL TOXICOL CHEM | 1 | 12% | 3.1% | 10 |
| 7 | MICRONUTR UNIT | 1 | 40% | 0.6% | 2 |
| 8 | CIENCIAS QUIMICAS FARMACEUT | 1 | 50% | 0.3% | 1 |
| 9 | ENVIRONM IND HYG | 1 | 50% | 0.3% | 1 |
| 10 | EXPOSURE ASSESSMENT COORDINAT POLICY | 1 | 50% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000201325 | GLUTATHIONE TRANSFERASE//GLUTATHIONE S TRANSFERASE//GLUTATHIONE S TRANSFERASE PI |
| 2 | 0.0000188711 | MPGES 1//PROSTAGLANDIN E SYNTHASE//MPGES 2 |
| 3 | 0.0000139779 | 1 HYDROXYETHYL RADICALS//AICA P//ETHANOL INDUCED HEPATOCELLULAR INJURY |
| 4 | 0.0000105965 | SALMONELLA ZENTRALE//URIDINE DIPHOSPHOGLUCURONYL TRANSFERASE//BUNDES INFEKTIONSKRANKHEITEN NICHT UBERTRAG |
| 5 | 0.0000098692 | CHEM DEV CHEM DEV OPERAT//WURTZ CONDENSATION//HEXAFLUOROBUTADIENE |
| 6 | 0.0000092133 | ENGN REACT CHEM//OXIDATION OF DNA//ALPHA TOCOPHERYL QUINONE |
| 7 | 0.0000087748 | FITTING OF NONLINEAR EQUATIONS//INSERMU392//GRP ENZYME BIOTECHNOL GENET ENGN |
| 8 | 0.0000082182 | PHYSICAL AND CHEMICAL NOXAE//MURINE SPERMATOGENESIS//THENOYLTRIFLUORACETONE |
| 9 | 0.0000068763 | 5 LIPOXYGENASE//LEUKOTRIENE A4 HYDROLASE//5 LIPOXYGENASE ACTIVATING PROTEIN |
| 10 | 0.0000067360 | ECHINOISOFLAVANONE//SOPHORA CHRYSOPHYLLA SEEM//SOPHORAISOFLAVANONE D |