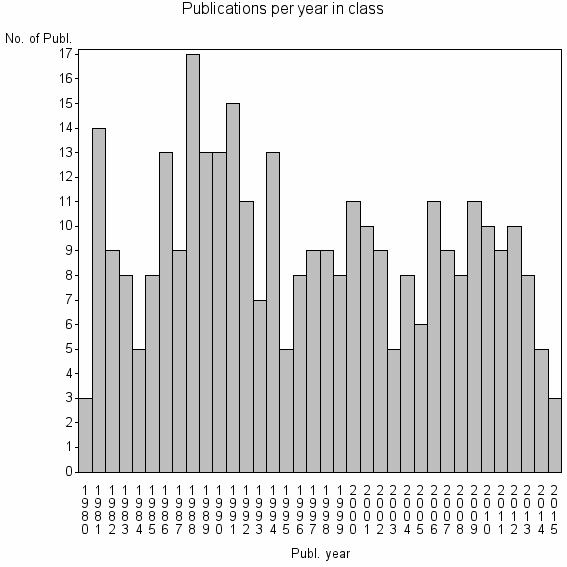
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 22293 | 330 | 38.2 | 76% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 302 | 17871 | TRANSLESION SYNTHESIS//DNA POLYMERASE//RECA PROTEIN |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | UVRB | Author keyword | 21 | 71% | 5% | 17 |
| 2 | UVRABC | Author keyword | 8 | 62% | 2% | 8 |
| 3 | UVRC | Author keyword | 5 | 63% | 2% | 5 |
| 4 | UVRA | Author keyword | 5 | 44% | 2% | 8 |
| 5 | ABC EXCINUCLEASE | Author keyword | 3 | 100% | 1% | 3 |
| 6 | DAMAGE RECOGNITION | Author keyword | 2 | 20% | 3% | 10 |
| 7 | UVRABC ENDONUCLEASE | Author keyword | 2 | 50% | 1% | 3 |
| 8 | MFD | Author keyword | 2 | 23% | 2% | 6 |
| 9 | BACTERIAL NUCLEOTIDE EXCISION REPAIR | Author keyword | 1 | 100% | 1% | 2 |
| 10 | UVRAB COMPLEX | Author keyword | 1 | 100% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | UVRB | 21 | 71% | 5% | 17 | Search UVRB | Search UVRB |
| 2 | UVRABC | 8 | 62% | 2% | 8 | Search UVRABC | Search UVRABC |
| 3 | UVRC | 5 | 63% | 2% | 5 | Search UVRC | Search UVRC |
| 4 | UVRA | 5 | 44% | 2% | 8 | Search UVRA | Search UVRA |
| 5 | ABC EXCINUCLEASE | 3 | 100% | 1% | 3 | Search ABC+EXCINUCLEASE | Search ABC+EXCINUCLEASE |
| 6 | DAMAGE RECOGNITION | 2 | 20% | 3% | 10 | Search DAMAGE+RECOGNITION | Search DAMAGE+RECOGNITION |
| 7 | UVRABC ENDONUCLEASE | 2 | 50% | 1% | 3 | Search UVRABC+ENDONUCLEASE | Search UVRABC+ENDONUCLEASE |
| 8 | MFD | 2 | 23% | 2% | 6 | Search MFD | Search MFD |
| 9 | BACTERIAL NUCLEOTIDE EXCISION REPAIR | 1 | 100% | 1% | 2 | Search BACTERIAL+NUCLEOTIDE+EXCISION+REPAIR | Search BACTERIAL+NUCLEOTIDE+EXCISION+REPAIR |
| 10 | UVRAB COMPLEX | 1 | 100% | 1% | 2 | Search UVRAB+COMPLEX | Search UVRAB+COMPLEX |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PREINCISION COMPLEX | 112 | 97% | 10% | 32 |
| 2 | ABC EXCINUCLEASE | 72 | 56% | 27% | 89 |
| 3 | ESCHERICHIA COLI UVRA | 34 | 79% | 7% | 22 |
| 4 | 5 INCISION | 33 | 75% | 7% | 24 |
| 5 | UVRABC ENDONUCLEASE | 17 | 59% | 6% | 19 |
| 6 | INCISION REACTION | 14 | 100% | 2% | 7 |
| 7 | ESCHERICHIA COLI UVRAB | 13 | 80% | 2% | 8 |
| 8 | UVRB | 12 | 52% | 5% | 16 |
| 9 | FACTOR MFD | 11 | 78% | 2% | 7 |
| 10 | ESCHERICHIA COLI UVRB | 10 | 58% | 3% | 11 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Prokaryotic nucleotide excision repair: The UvrABC system | 2006 | 145 | 207 | 57% |
| 'Close-fitting sleeves': DNA damage recognition by the UvrABC nuclease system | 2005 | 76 | 168 | 56% |
| The molecular mechanism of transcription-coupled DNA repair | 2007 | 37 | 41 | 46% |
| Repair of UV damage in bacteria | 2008 | 56 | 81 | 36% |
| Transcription-Coupled DNA Repair in Prokaryotes | 2012 | 14 | 62 | 42% |
| NUCLEOTIDE EXCISION-REPAIR, A TRACKING MECHANISM IN SEARCH OF DAMAGE | 1993 | 57 | 35 | 74% |
| Surviving the sun: Repair and bypass of DNA UV lesions | 2011 | 16 | 68 | 35% |
| Rethinking transcription coupled DNA repair | 2015 | 1 | 52 | 31% |
| MECHANISM OF ACTION OF THE ESCHERICHIA-COLI UVRABC NUCLEASE - CLUES TO THE DAMAGE RECOGNITION PROBLEM | 1993 | 68 | 44 | 52% |
| NUCLEOTIDE EXCISION REPAIR IN ESCHERICHIA-COLI | 1990 | 373 | 218 | 30% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DNA PROT INTERACT UNIT | 1 | 11% | 2.4% | 8 |
| 2 | CHEM MICROBIOL BIOCHEM BIOL | 1 | 50% | 0.3% | 1 |
| 3 | DNA PROTEIN INTERACT UNIT | 1 | 50% | 0.3% | 1 |
| 4 | FARMACOS MICROBIOL IND AVALIACAO GENOT | 1 | 50% | 0.3% | 1 |
| 5 | PX THER EUT | 1 | 50% | 0.3% | 1 |
| 6 | UNIDADE GENOM | 1 | 50% | 0.3% | 1 |
| 7 | RADIOBIOL MOL | 0 | 11% | 1.2% | 4 |
| 8 | LAWRENCE BERKELEY CHEM | 0 | 33% | 0.3% | 1 |
| 9 | LAWRENCE BERKELEY CHEM BIODYNAM | 0 | 33% | 0.3% | 1 |
| 10 | O ANAL CLIN TOXICOL | 0 | 33% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000208490 | XERODERMA PIGMENTOSUM//COCKAYNE SYNDROME//NUCLEOTIDE EXCISION REPAIR |
| 2 | 0.0000202502 | SOS MUTAGENESIS//UMUDC//LEXA REPRESSOR |
| 3 | 0.0000164749 | OBSTET GYNECOLSEALY STRUCT BIOL//PRIMASE//HELICASE |
| 4 | 0.0000125470 | PHOTOLYASE//DNA PHOTOLYASE//PHOTOLYASES |
| 5 | 0.0000116728 | NITRENIUM IONS//NITRENIUM//N 2 ACETYLAMINOFLUORENE |
| 6 | 0.0000086930 | FUROCOUMARINS//DNA PHOTOBINDING//PHOTOBIOLOGICAL ACTIVITY |
| 7 | 0.0000081415 | HOLLIDAY JUNCTION//RECBCD ENZYME//RECBCD |
| 8 | 0.0000059751 | DEINOCOCCUS RADIODURANS//DEINOCOCCUS//DEINOCOCCACEAE |
| 9 | 0.0000058707 | DNA GLYCOSYLASE//8 OXOGUANINE//BASE EXCISION REPAIR |
| 10 | 0.0000058241 | ENDONUCLEASE V//S CAROLINA EXPT STN//OXANINE |