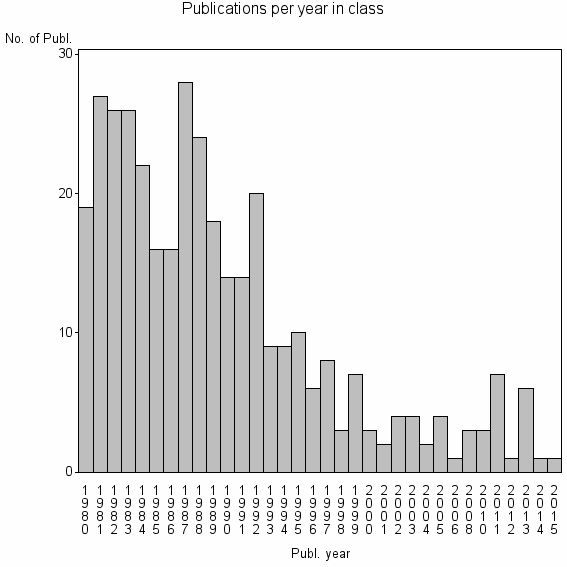
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 21319 | 364 | 30.7 | 43% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 3084 | 1884 | MYXOMYCETES//PHYSARUM POLYCEPHALUM//SLIME MOLDS |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | MAGDEBURG SYST BIOL MACS | Address | 9 | 67% | 2% | 8 |
| 2 | PHYSARUM POLYCEPHALUM | Author keyword | 7 | 16% | 11% | 41 |
| 3 | MAGDEBURG SYST BIOL | Address | 5 | 63% | 1% | 5 |
| 4 | P POLYCEPHALUM | Author keyword | 3 | 50% | 1% | 4 |
| 5 | PHYSARUM | Author keyword | 2 | 16% | 4% | 13 |
| 6 | RAP GENE | Author keyword | 2 | 67% | 1% | 2 |
| 7 | MITOCHONDRIAL PLASMID | Author keyword | 2 | 22% | 2% | 7 |
| 8 | BIOCHEM EXPTL CANC | Address | 1 | 100% | 1% | 2 |
| 9 | LEHRSTUHL REGULAT BIOL | Address | 1 | 100% | 1% | 2 |
| 10 | PLASMODIUM DEVELOPMENT | Author keyword | 1 | 100% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PHYSARUM POLYCEPHALUM | 7 | 16% | 11% | 41 | Search PHYSARUM+POLYCEPHALUM | Search PHYSARUM+POLYCEPHALUM |
| 2 | P POLYCEPHALUM | 3 | 50% | 1% | 4 | Search P+POLYCEPHALUM | Search P+POLYCEPHALUM |
| 3 | PHYSARUM | 2 | 16% | 4% | 13 | Search PHYSARUM | Search PHYSARUM |
| 4 | RAP GENE | 2 | 67% | 1% | 2 | Search RAP+GENE | Search RAP+GENE |
| 5 | MITOCHONDRIAL PLASMID | 2 | 22% | 2% | 7 | Search MITOCHONDRIAL+PLASMID | Search MITOCHONDRIAL+PLASMID |
| 6 | PLASMODIUM DEVELOPMENT | 1 | 100% | 1% | 2 | Search PLASMODIUM+DEVELOPMENT | Search PLASMODIUM+DEVELOPMENT |
| 7 | JELLY ROLL MOTIF | 1 | 50% | 0% | 1 | Search JELLY+ROLL+MOTIF | Search JELLY+ROLL+MOTIF |
| 8 | LYSINE RICH DOMAIN | 1 | 50% | 0% | 1 | Search LYSINE+RICH+DOMAIN | Search LYSINE+RICH+DOMAIN |
| 9 | MITOCHONDRIAL REPLICON CLUSTER | 1 | 50% | 0% | 1 | Search MITOCHONDRIAL+REPLICON+CLUSTER | Search MITOCHONDRIAL+REPLICON+CLUSTER |
| 10 | MODELING BIOLOGICAL SYSTEMS | 1 | 50% | 0% | 1 | Search MODELING+BIOLOGICAL+SYSTEMS | Search MODELING+BIOLOGICAL+SYSTEMS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RESOLVED SOMATIC COMPLEMENTATION | 17 | 100% | 2% | 8 |
| 2 | VARIABLE PATHWAYS | 15 | 88% | 2% | 7 |
| 3 | PLASMODIA | 4 | 30% | 3% | 10 |
| 4 | PLASMID MF | 2 | 67% | 1% | 2 |
| 5 | POLYCEPHALUM | 2 | 13% | 4% | 14 |
| 6 | PLASMODIAL TRANSITION | 1 | 100% | 1% | 2 |
| 7 | MYXOMYCETE | 1 | 20% | 1% | 4 |
| 8 | UNIPARENTAL INHERITANCE | 1 | 11% | 2% | 7 |
| 9 | MYXOMYCETE PHYSARUM POLYCEPHALUM | 1 | 33% | 1% | 2 |
| 10 | SANDWICHED PLASMODIA | 1 | 50% | 0% | 1 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| PLASMODIUM DEVELOPMENT IN THE MYXOMYCETE PHYSARUM-POLYCEPHALUM - GENETIC-CONTROL AND CELLULAR EVENTS | 1995 | 13 | 52 | 87% |
| PATTERNS OF INHERITANCE, DEVELOPMENT AND THE MITOTIC-CYCLE IN THE PROTIST PHYSARUM-POLYCEPHALUM | 1993 | 44 | 175 | 50% |
| Detecting functional interactions in a gene and signaling network by time-resolved somatic complementation analysis | 2003 | 4 | 36 | 69% |
| Mitochondrial fusion promoting plasmid | 1997 | 2 | 38 | 39% |
| Sexuality of mitochondria: Fusion, recombination, and plasmids | 1995 | 20 | 110 | 16% |
| PERIODIC EVENTS AND CELL-CYCLE REGULATION IN THE PLASMODIA OF PHYSARUM-POLYCEPHALUM | 1995 | 1 | 153 | 39% |
| THE TUBULIN AND HISTONE GENES OF PHYSARUM-POLYCEPHALUM - MODELS FOR CELL CYCLE-REGULATED GENE-EXPRESSION | 1986 | 7 | 13 | 62% |
| SURFACE CHANGES DURING GROWTH AND DEVELOPMENT OF SLIME-MOLDS | 1994 | 0 | 8 | 25% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MAGDEBURG SYST BIOL MACS | 9 | 67% | 2.2% | 8 |
| 2 | MAGDEBURG SYST BIOL | 5 | 63% | 1.4% | 5 |
| 3 | BIOCHEM EXPTL CANC | 1 | 100% | 0.5% | 2 |
| 4 | LEHRSTUHL REGULAT BIOL | 1 | 100% | 0.5% | 2 |
| 5 | PLANT LIFE SYST | 1 | 40% | 0.5% | 2 |
| 6 | SEKT BIOL CHEM | 1 | 50% | 0.3% | 1 |
| 7 | MATH MODELLING MODEMAT | 0 | 25% | 0.3% | 1 |
| 8 | EXPTL CANC | 0 | 17% | 0.3% | 1 |
| 9 | MAX PLANCK DYNAM COMPLEX TECH SYST | 0 | 14% | 0.3% | 1 |
| 10 | ZELLBIOL BIOCHEM BIOTECHNOL | 0 | 14% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000234302 | UNCONVENT COMP//PHYSARUM POLYCEPHALUM//PHYSARUM |
| 2 | 0.0000167460 | MYXOMYCETES//SLIME MOLDS//DICTYOSTELIDS |
| 3 | 0.0000158841 | TUBULIN ISOTYPES//TYROSINATION//DETYROSINATION |
| 4 | 0.0000104812 | POLYMALIC ACID//POLYMALATASE//UMR C7581 |
| 5 | 0.0000103471 | PHOSPHATIDYLGLUCOSIDE//CHOLESTEROL ALPHA GLUCOSYLTRANSFERASE//EURYALE FEROX |
| 6 | 0.0000087578 | PLASTID INHERITANCE//PLASTOME GENOME INCOMPATIBILITY//BIPARENTAL INHERITANCE |
| 7 | 0.0000084526 | CHLAMYDOMONAS MONOICA//CELL CYCLES//CELL CYCLES BIOTECHNOL ALGAE |
| 8 | 0.0000072875 | ULTRAVIOLET MICROBEAM//CRANE FLY SPERMATOCYTES//SPINDLE FIBRES |
| 9 | 0.0000061274 | THYMIDINE KINASE 1//SERUM THYMIDINE KINASE 1//DNA SYNTHESIZING ENZYMES |
| 10 | 0.0000055715 | PODOSPORA ANSERINA//LINEAR MITOCHONDRIAL PLASMID//KALILO |