Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
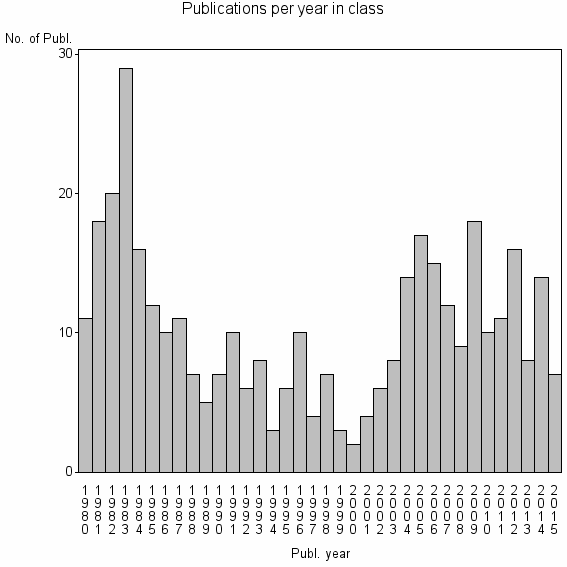
| 21057 | 374 | 33.4 | 63% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2787 | 2596 | DIHYDRODIPICOLINATE SYNTHASE//ASPARTATE KINASE//METHIONINE DEPENDENCE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | METHYLTHIOADENOSINE PHOSPHORYLASE | Author keyword | 15 | 71% | 3% | 12 |
| 2 | MTAP | Author keyword | 12 | 54% | 4% | 15 |
| 3 | L ALANOSINE | Author keyword | 9 | 83% | 1% | 5 |
| 4 | METHIONINE SALVAGE PATHWAY | Author keyword | 8 | 56% | 3% | 10 |
| 5 | METHYLTHIOADENOSINE | Author keyword | 7 | 56% | 2% | 9 |
| 6 | HINEKA | Author keyword | 6 | 100% | 1% | 4 |
| 7 | ALPHA KETOGLUTARAMATE | Author keyword | 5 | 63% | 1% | 5 |
| 8 | OMEGA AMIDASE | Author keyword | 4 | 56% | 1% | 5 |
| 9 | 1 2 DIHYDROXY 5 METHYLSULFINYLPENTAN 3 ONE | Author keyword | 3 | 100% | 1% | 3 |
| 10 | 5 METHYLTHIOADENOSINE NUCLEOSIDASE | Author keyword | 3 | 100% | 1% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | METHYLTHIOADENOSINE PHOSPHORYLASE | 15 | 71% | 3% | 12 | Search METHYLTHIOADENOSINE+PHOSPHORYLASE | Search METHYLTHIOADENOSINE+PHOSPHORYLASE |
| 2 | MTAP | 12 | 54% | 4% | 15 | Search MTAP | Search MTAP |
| 3 | L ALANOSINE | 9 | 83% | 1% | 5 | Search L+ALANOSINE | Search L+ALANOSINE |
| 4 | METHIONINE SALVAGE PATHWAY | 8 | 56% | 3% | 10 | Search METHIONINE+SALVAGE+PATHWAY | Search METHIONINE+SALVAGE+PATHWAY |
| 5 | METHYLTHIOADENOSINE | 7 | 56% | 2% | 9 | Search METHYLTHIOADENOSINE | Search METHYLTHIOADENOSINE |
| 6 | HINEKA | 6 | 100% | 1% | 4 | Search HINEKA | Search HINEKA |
| 7 | ALPHA KETOGLUTARAMATE | 5 | 63% | 1% | 5 | Search ALPHA+KETOGLUTARAMATE | Search ALPHA+KETOGLUTARAMATE |
| 8 | OMEGA AMIDASE | 4 | 56% | 1% | 5 | Search OMEGA+AMIDASE | Search OMEGA+AMIDASE |
| 9 | 1 2 DIHYDROXY 5 METHYLSULFINYLPENTAN 3 ONE | 3 | 100% | 1% | 3 | Search 1+2+DIHYDROXY+5+METHYLSULFINYLPENTAN+3+ONE | Search 1+2+DIHYDROXY+5+METHYLSULFINYLPENTAN+3+ONE |
| 10 | 5 METHYLTHIOADENOSINE NUCLEOSIDASE | 3 | 100% | 1% | 3 | Search 5++METHYLTHIOADENOSINE+NUCLEOSIDASE | Search 5++METHYLTHIOADENOSINE+NUCLEOSIDASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PURINE METABOLIC ENZYME | 53 | 95% | 5% | 18 |
| 2 | 5 METHYLTHIOADENOSINE | 34 | 61% | 10% | 36 |
| 3 | 5 METHYLTHIORIBOSE KINASE | 29 | 88% | 4% | 14 |
| 4 | MTAP | 27 | 92% | 3% | 11 |
| 5 | CODELETION | 15 | 71% | 3% | 12 |
| 6 | MTA ADOHCY NUCLEOSIDASE | 11 | 100% | 2% | 6 |
| 7 | 5 METHYLTHIORIBOSE | 11 | 69% | 2% | 9 |
| 8 | 5 DEOXY 5 METHYLTHIOADENOSINE | 8 | 52% | 3% | 11 |
| 9 | L ALANOSINE NSC 153353 | 8 | 100% | 1% | 5 |
| 10 | METHYLTHIOADENOSINE PHOSPHORYLASE | 8 | 28% | 6% | 23 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Metabolic Characteristics and Importance of the Universal Methionine Salvage Pathway Recycling Methionine from 5 '-Methylthioadenosine | 2009 | 49 | 89 | 62% |
| Targeting tumors that lack methylthioadenosine phosphorylase (MTAP) activity Current strategies | 2011 | 15 | 45 | 80% |
| Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis | 2013 | 24 | 79 | 13% |
| Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism | 2011 | 48 | 120 | 17% |
| The logic of metabolism and its fuzzy consequences | 2014 | 2 | 44 | 9% |
| METHYLTHIOADENOSINE | 1983 | 66 | 47 | 53% |
| TRENDS IN THE BIOCHEMICAL PHARMACOLOGY OF 5'-DEOXY-5'-METHYLTHIOADENOSINE | 1982 | 187 | 27 | 59% |
| 5'-METHYLTHIOADENOSINE PHOSPHORYLASE-DEFICIENCY IN MALIGNANT-CELLS - RECESSIVE EXPRESSION OF THE DEFECTIVE PHENOTYPE IN INTRASPECIES (MOUSE X MOUSE) HYBRIDS | 1984 | 1 | 3 | 100% |
| 5-METHYLTHIORIBOSE KINASE (ENTEROBACTER-AEROGENES) | 1983 | 5 | 5 | 100% |
| 5'-METHYLTHIOADENOSINE NUCLEOSIDASE (LUPINUS-LUTEUS SEEDS) | 1983 | 0 | 1 | 100% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CLIN GENET UNIV REG S | 1 | 50% | 0.3% | 1 |
| 2 | TECHNOL PK BIZKAIA | 1 | 50% | 0.3% | 1 |
| 3 | THERAGENETEX BIO | 1 | 50% | 0.3% | 1 |
| 4 | IND MACROMOL CRYSTALLOG ASSOC | 0 | 33% | 0.3% | 1 |
| 5 | FIMM TECHNOL | 0 | 25% | 0.3% | 1 |
| 6 | METABOLOM UNIT | 0 | 20% | 0.3% | 1 |
| 7 | ONCOL BREAST SURG | 0 | 20% | 0.3% | 1 |
| 8 | HEIDELBERG PLANT SCI HIP | 0 | 17% | 0.3% | 1 |
| 9 | UMR 6519 CNRS | 0 | 17% | 0.3% | 1 |
| 10 | MIDW PROTEOME | 0 | 14% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000203063 | METHIONINE DEPENDENCE//METHIONINASE//L METHIONINE GAMMA LYASE |
| 2 | 0.0000197804 | PURINE NUCLEOSIDE PHOSPHORYLASE//NUCLEOSIDE PHOSPHORYLASES//CARBOHYDRATE CHEM TEAM |
| 3 | 0.0000161041 | QUERCETINASE//BAI LIGANDS//QUERCETIN 2 3 DIOXYGENASE |
| 4 | 0.0000118220 | APRT DEFICIENCY//ADENINE PHOSPHORIBOSYLTRANSFERASE DEFICIENCY//2 8 DIHYDROXYADENINE |
| 5 | 0.0000114669 | S ADENOSYLMETHIONINE DECARBOXYLASE//CATIONIC DIAMIDINES//DFMO RESISTANCE |
| 6 | 0.0000112739 | METHIONINE ADENOSYLTRANSFERASE//S ADENOSYLMETHIONINE//GLYCINE N METHYLTRANSFERASE |
| 7 | 0.0000095884 | P14ARF//P16//P16INK4A |
| 8 | 0.0000092935 | CARBOCYCLIC NUCLEOSIDES//S ADENOSYLHOMOCYSTEINE HYDROLASE//CARBOCYCLIC NUCLEOSIDE |
| 9 | 0.0000071825 | URINARY NUCLEOSIDES//URINARY MODIFIED NUCLEOSIDES//MODIFIED NUCLEOSIDES |
| 10 | 0.0000068676 | POLYAMINES//SPERMIDINE SPERMINE N 1 ACETYLTRANSFERASE//SPERMINE OXIDASE |