Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
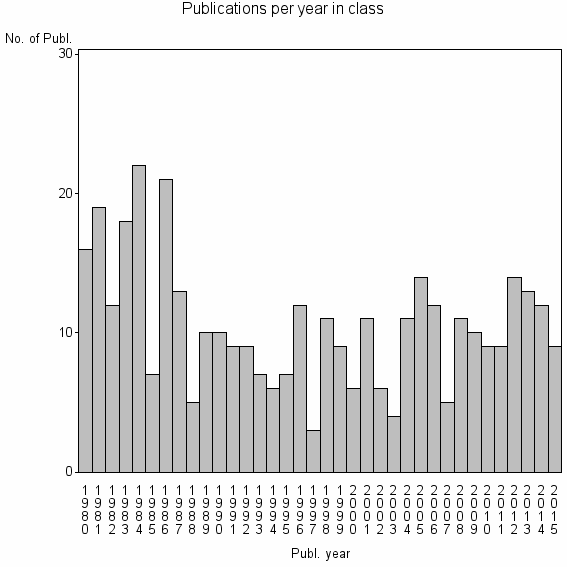
| 20863 | 382 | 33.8 | 57% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RIBOSOME MODULATION FACTOR | Author keyword | 9 | 83% | 1% | 5 |
| 2 | AMINE PHARMA | Address | 6 | 34% | 4% | 14 |
| 3 | POLYAMINE MODULON | Author keyword | 6 | 100% | 1% | 4 |
| 4 | 100S RIBOSOME | Author keyword | 4 | 75% | 1% | 3 |
| 5 | NCI H716 | Author keyword | 3 | 100% | 1% | 3 |
| 6 | POLYAMINE BINDING | Author keyword | 2 | 50% | 1% | 3 |
| 7 | POTE | Author keyword | 1 | 100% | 1% | 2 |
| 8 | PUTRESCINE AMINOTRANSFERASE | Author keyword | 1 | 100% | 1% | 2 |
| 9 | PUTRESCINE DEGRADATION | Author keyword | 1 | 100% | 1% | 2 |
| 10 | RIBOSOME DIMERISATION | Author keyword | 1 | 100% | 1% | 2 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RIBOSOME MODULATION FACTOR | 9 | 83% | 1% | 5 | Search RIBOSOME+MODULATION+FACTOR | Search RIBOSOME+MODULATION+FACTOR |
| 2 | POLYAMINE MODULON | 6 | 100% | 1% | 4 | Search POLYAMINE+MODULON | Search POLYAMINE+MODULON |
| 3 | 100S RIBOSOME | 4 | 75% | 1% | 3 | Search 100S+RIBOSOME | Search 100S+RIBOSOME |
| 4 | NCI H716 | 3 | 100% | 1% | 3 | Search NCI+H716 | Search NCI+H716 |
| 5 | POLYAMINE BINDING | 2 | 50% | 1% | 3 | Search POLYAMINE+BINDING | Search POLYAMINE+BINDING |
| 6 | POTE | 1 | 100% | 1% | 2 | Search POTE | Search POTE |
| 7 | PUTRESCINE AMINOTRANSFERASE | 1 | 100% | 1% | 2 | Search PUTRESCINE+AMINOTRANSFERASE | Search PUTRESCINE+AMINOTRANSFERASE |
| 8 | PUTRESCINE DEGRADATION | 1 | 100% | 1% | 2 | Search PUTRESCINE+DEGRADATION | Search PUTRESCINE+DEGRADATION |
| 9 | RIBOSOME DIMERISATION | 1 | 100% | 1% | 2 | Search RIBOSOME+DIMERISATION | Search RIBOSOME+DIMERISATION |
| 10 | RIBOSOME HIBERNATION | 1 | 100% | 1% | 2 | Search RIBOSOME+HIBERNATION | Search RIBOSOME+HIBERNATION |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PUTRESCINE TRANSPORT SYSTEM | 23 | 79% | 4% | 15 |
| 2 | PREFERENTIAL UPTAKE SYSTEM | 19 | 68% | 4% | 17 |
| 3 | 100S RIBOSOME | 12 | 86% | 2% | 6 |
| 4 | MODULATION FACTOR RMF | 10 | 73% | 2% | 8 |
| 5 | SELECTIVE STRUCTURAL CHANGE | 8 | 100% | 1% | 5 |
| 6 | OLIGOPEPTIDE BINDING PROTEIN | 7 | 36% | 4% | 16 |
| 7 | YFIA | 6 | 80% | 1% | 4 |
| 8 | MODULATION FACTOR | 6 | 41% | 3% | 11 |
| 9 | BULGED OUT REGION | 6 | 100% | 1% | 4 |
| 10 | 30 S RIBOSOMAL SUBUNITS | 5 | 60% | 2% | 6 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Characteristics of cellular polyamine transport in prokaryotes and eukaryotes | 2010 | 48 | 65 | 48% |
| Polyamine modulon in Escherichia coli: Genes involved in the stimulation of cell growth by polyamines | 2006 | 69 | 31 | 48% |
| The 100S ribosome: ribosomal hibernation induced by stress | 2014 | 2 | 46 | 54% |
| MECHANISM OF BACTERICIDAL ACTION OF AMINOGLYCOSIDES | 1987 | 268 | 19 | 74% |
| Growth phase coupled modulation of Escherichia coli ribosomes | 1998 | 53 | 16 | 44% |
| Structure and function of polyamine-amino acid antiporters CadB and PotE in Escherichia coli | 2012 | 4 | 26 | 58% |
| Polyamine transport in bacteria and yeast | 1999 | 134 | 85 | 26% |
| A multifaceted role for polyamines in bacterial pathogens | 2008 | 65 | 112 | 19% |
| BACTERIAL UPTAKE OF AMINOGLYCOSIDE ANTIBIOTICS | 1987 | 187 | 77 | 51% |
| Physiological functions of polyamines and regulation of polyamine content in cells | 2006 | 6 | 61 | 49% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | AMINE PHARMA | 6 | 34% | 3.7% | 14 |
| 2 | CHUOUKU | 1 | 50% | 0.3% | 1 |
| 3 | GRP MOL STRUCT BIOPHYS | 0 | 25% | 0.3% | 1 |
| 4 | LOWER MALL STN | 0 | 12% | 0.5% | 2 |
| 5 | CLIN ANALYT BIOCHEM | 0 | 20% | 0.3% | 1 |
| 6 | SECT BACTERIOL AQUAT TERR | 0 | 20% | 0.3% | 1 |
| 7 | INTEGRATED METABOL GRP | 0 | 10% | 0.5% | 2 |
| 8 | PLASMA MEMBRANE NUCL SIGNALLING | 0 | 17% | 0.3% | 1 |
| 9 | UMEA MICROBIAL UCMR | 0 | 14% | 0.3% | 1 |
| 10 | GENET MICROORGANISMOS | 0 | 11% | 0.3% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000156200 | PL MICROBIOLAOBA KU//CALDOPENTAMINE//HOMOSPERMIDINE |
| 2 | 0.0000126919 | POLYAMINES//SPERMIDINE SPERMINE N 1 ACETYLTRANSFERASE//SPERMINE OXIDASE |
| 3 | 0.0000108005 | PINEAL GLAND PEPTIDES//SUBSTRAIN DIFFERENCE//CAROLINA ENVIRONM BIOINFORMAT |
| 4 | 0.0000107889 | S ADENOSYLMETHIONINE DECARBOXYLASE//CATIONIC DIAMIDINES//DFMO RESISTANCE |
| 5 | 0.0000099559 | PPGPP//STRINGENT RESPONSE//RPOS |
| 6 | 0.0000095395 | AMINOGLYCOSIDE//AMINOGLYCOSIDES//RNA RECOGNITION |
| 7 | 0.0000092754 | PHARMACEUT SCI BUNKYO MACHI 114//SELECTED ANTIBODY ENZYME IMMUNOASSAY//CELL BIOL IBHV |
| 8 | 0.0000084395 | AMMONIA SELECTIVE ELECTRODE//BOT CROP PHYSIOL//ELECTROPHYSICAL |
| 9 | 0.0000081063 | POSTANTIBIOTIC EFFECT//POSTANTIBIOTIC SUB MIC EFFECT//POST ANTIBIOTIC EFFECT |
| 10 | 0.0000076435 | ANTIZYME INHIBITOR//ANTIZYME//ORNITHINE DECARBOXYLASE |