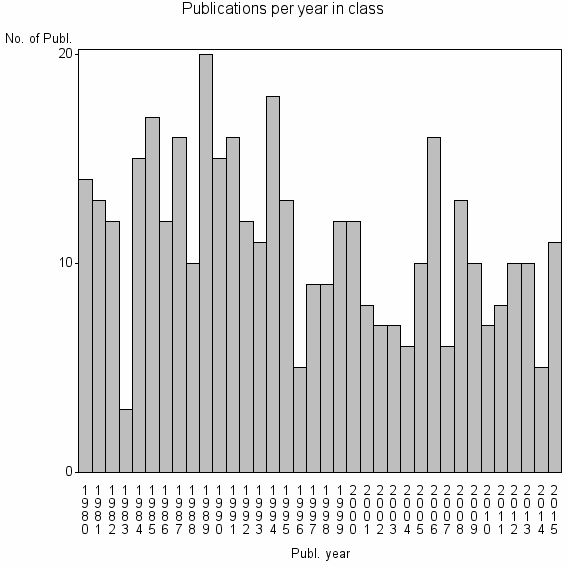
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 20457 | 398 | 33.6 | 58% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | TRANSCRIPTION FACTOR TNRA | Author keyword | 6 | 100% | 1% | 4 |
| 2 | GLNR | Author keyword | 2 | 33% | 1% | 4 |
| 3 | BECKMAN COULTER | Author keyword | 1 | 50% | 1% | 2 |
| 4 | GLUTAMINE SYNTHETASE TYPE III | Author keyword | 1 | 100% | 1% | 2 |
| 5 | GSIII | Author keyword | 1 | 100% | 1% | 2 |
| 6 | POLY L GLUTAMINE GLUTAMATE | Author keyword | 1 | 100% | 1% | 2 |
| 7 | RAP RT PCR | Author keyword | 1 | 50% | 1% | 2 |
| 8 | TNRA | Author keyword | 1 | 100% | 1% | 2 |
| 9 | CUII AMINO ACID COMPLEXES | Author keyword | 1 | 50% | 0% | 1 |
| 10 | ECL WESTERN IMMUNOBLOTTING | Author keyword | 1 | 50% | 0% | 1 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | TRANSCRIPTION FACTOR TNRA | 6 | 100% | 1% | 4 | Search TRANSCRIPTION+FACTOR+TNRA | Search TRANSCRIPTION+FACTOR+TNRA |
| 2 | GLNR | 2 | 33% | 1% | 4 | Search GLNR | Search GLNR |
| 3 | BECKMAN COULTER | 1 | 50% | 1% | 2 | Search BECKMAN+COULTER | Search BECKMAN+COULTER |
| 4 | GLUTAMINE SYNTHETASE TYPE III | 1 | 100% | 1% | 2 | Search GLUTAMINE+SYNTHETASE+TYPE+III | Search GLUTAMINE+SYNTHETASE+TYPE+III |
| 5 | GSIII | 1 | 100% | 1% | 2 | Search GSIII | Search GSIII |
| 6 | POLY L GLUTAMINE GLUTAMATE | 1 | 100% | 1% | 2 | Search POLY+L+GLUTAMINE+GLUTAMATE | Search POLY+L+GLUTAMINE+GLUTAMATE |
| 7 | RAP RT PCR | 1 | 50% | 1% | 2 | Search RAP+RT+PCR | Search RAP+RT+PCR |
| 8 | TNRA | 1 | 100% | 1% | 2 | Search TNRA | Search TNRA |
| 9 | CUII AMINO ACID COMPLEXES | 1 | 50% | 0% | 1 | Search CUII+AMINO+ACID+COMPLEXES | Search CUII+AMINO+ACID+COMPLEXES |
| 10 | ECL WESTERN IMMUNOBLOTTING | 1 | 50% | 0% | 1 | Search ECL+WESTERN+IMMUNOBLOTTING | Search ECL+WESTERN+IMMUNOBLOTTING |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | TRANSCRIPTION FACTOR TNRA | 22 | 81% | 3% | 13 |
| 2 | GLUTAMINE SYNTHETASE MUTANTS | 17 | 100% | 2% | 8 |
| 3 | TNRA | 11 | 60% | 3% | 12 |
| 4 | GLNA GENE | 11 | 69% | 2% | 9 |
| 5 | GAMMA SUBSTITUTED PHOSPHINOTHRICINS | 7 | 53% | 2% | 9 |
| 6 | GLNRA OPERON | 6 | 100% | 1% | 4 |
| 7 | PHOSPHINOTHRICYL ALANYL ALANINE | 5 | 60% | 2% | 6 |
| 8 | GENE GLNA | 4 | 75% | 1% | 3 |
| 9 | GLNA1 | 4 | 75% | 1% | 3 |
| 10 | L METHIONINE SULFOXIMINE | 4 | 56% | 1% | 5 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Control of glutamate homeostasis in Bacillus subtilis: a complex interplay between ammonium assimilation, glutamate biosynthesis and degradation | 2012 | 22 | 93 | 47% |
| Structure-function relationships of glutamine synthetases | 2000 | 163 | 65 | 60% |
| Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! | 1999 | 163 | 47 | 43% |
| Common patterns - unique features: nitrogen metabolism and regulation in Gram-positive bacteria | 2010 | 24 | 150 | 26% |
| Inhibitors of glutamine synthetase and their potential application in medicine | 2008 | 14 | 70 | 43% |
| Inhibition of Glutamine Synthetase: A Potential Drug Target in Mycobacterium tuberculosis | 2014 | 0 | 40 | 63% |
| Regulation of nitrogen metabolism in Mycobacterium tuberculosis: A comparison with mechanisms in Corynebacterium glutamicum and Streptomyces coelicolor | 2008 | 11 | 43 | 33% |
| RECENT DEVELOPMENTS ON THE REGULATION AND STRUCTURE OF GLUTAMINE-SYNTHETASE ENZYMES FROM SELECTED BACTERIAL GROUPS | 1993 | 29 | 56 | 50% |
| REGULATION OF ESCHERICHIA-COLI GLUTAMINE-SYNTHETASE | 1989 | 70 | 51 | 37% |
| Advances in the enzymology of glutamine synthesis | 1998 | 23 | 24 | 17% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MICROBIOM | 1 | 50% | 0.3% | 1 |
| 2 | SCP BIOL | 0 | 33% | 0.3% | 1 |
| 3 | TSU INFECT DIS | 0 | 33% | 0.3% | 1 |
| 4 | US MRC MOL CELLULAR BIOL HLTH SCI | 0 | 33% | 0.3% | 1 |
| 5 | BIOTECHNOL NANOMED SECT | 0 | 25% | 0.3% | 1 |
| 6 | INFORMAT TRANSMISS PROB | 0 | 25% | 0.3% | 1 |
| 7 | BIOCHEM BIOL CHEM | 0 | 20% | 0.3% | 1 |
| 8 | CIC ISLA CARTUJA | 0 | 20% | 0.3% | 1 |
| 9 | FB CHEM BIOCHEM | 0 | 20% | 0.3% | 1 |
| 10 | SECT PROT CHEM | 0 | 10% | 0.5% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000230664 | NIFA PROTEIN//SIGMA54//NIFA |
| 2 | 0.0000149379 | GLUTAMINE SYNTHETASE//GLUTAMATE SYNTHASE//CYTOSOLIC GLUTAMINE SYNTHETASE |
| 3 | 0.0000131319 | ABRB//SPO0A//TRANSITION STATE REGULATOR |
| 4 | 0.0000097639 | MOLEC GENE TECH//NOVEL POLYSACCHARIDE//BACILLUS ISOLATES |
| 5 | 0.0000082602 | GLUTAMATE DEHYDROGENASE//AMINO ACID DEHYDROGENASE//LEUCINE DEHYDROGENASE |
| 6 | 0.0000077706 | THEANINE//L THEANINE//COUPLED FERMENTATION WITH ENERGY TRANSFER |
| 7 | 0.0000066914 | SUCCINYL COA SYNTHETASE//OFF VP//CANADA GRP PROT STRUCT FUNCT |
| 8 | 0.0000060865 | NITROGEN METABOLITE REPRESSION//NMRA//FACB |
| 9 | 0.0000059236 | GENOME VECTOR//NIPPONKO KU//LONG ACCURATE PCR |
| 10 | 0.0000057339 | INOSITOL DEHYDROGENASE//INOSITOL CATABOLISM//MYO INOSITOL DEHYDROGENASE |