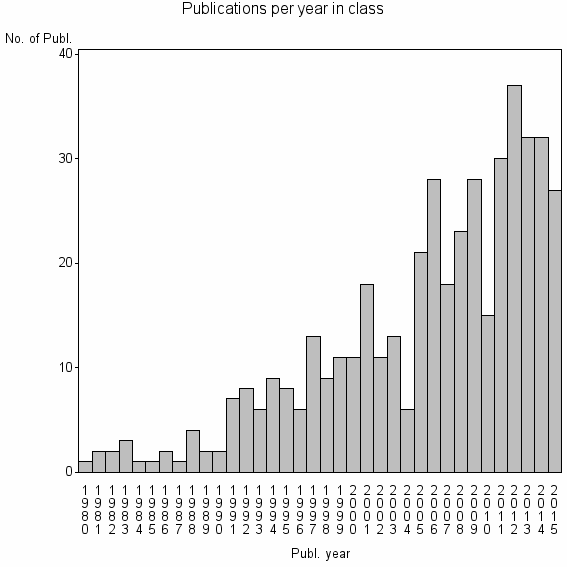
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 19254 | 448 | 46.1 | 79% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 3252 | 1456 | DISSOLVED DNA//NATURAL TRANSFORMATION//EDNA |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PROGRAM COMPUTAT SYST BIOL | Address | 3 | 35% | 1% | 6 |
| 2 | PERIODIC SELECTION | Author keyword | 1 | 31% | 1% | 4 |
| 3 | CHURINCE | Author keyword | 1 | 100% | 0% | 2 |
| 4 | PROKARYOTIC SPECIES | Author keyword | 1 | 50% | 0% | 2 |
| 5 | RECOMBINATION DETECTION | Author keyword | 1 | 33% | 1% | 3 |
| 6 | SPECIES DEFINITION | Author keyword | 1 | 21% | 1% | 4 |
| 7 | BIOINFORMAT EVOLUTIONARY GENOM GRP | Address | 1 | 33% | 0% | 2 |
| 8 | BACKBONE GENOME | Author keyword | 1 | 50% | 0% | 1 |
| 9 | BACTERIAL NOMENCLATURE | Author keyword | 1 | 50% | 0% | 1 |
| 10 | COMPUTAT GLOBAL SECUR | Address | 1 | 50% | 0% | 1 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PERIODIC SELECTION | 1 | 31% | 1% | 4 | Search PERIODIC+SELECTION | Search PERIODIC+SELECTION |
| 2 | CHURINCE | 1 | 100% | 0% | 2 | Search CHURINCE | Search CHURINCE |
| 3 | PROKARYOTIC SPECIES | 1 | 50% | 0% | 2 | Search PROKARYOTIC+SPECIES | Search PROKARYOTIC+SPECIES |
| 4 | RECOMBINATION DETECTION | 1 | 33% | 1% | 3 | Search RECOMBINATION+DETECTION | Search RECOMBINATION+DETECTION |
| 5 | SPECIES DEFINITION | 1 | 21% | 1% | 4 | Search SPECIES+DEFINITION | Search SPECIES+DEFINITION |
| 6 | BACKBONE GENOME | 1 | 50% | 0% | 1 | Search BACKBONE+GENOME | Search BACKBONE+GENOME |
| 7 | BACTERIAL NOMENCLATURE | 1 | 50% | 0% | 1 | Search BACTERIAL+NOMENCLATURE | Search BACTERIAL+NOMENCLATURE |
| 8 | DELTA TM | 1 | 50% | 0% | 1 | Search DELTA+TM | Search DELTA+TM |
| 9 | DOUBLE CROSSOVERS | 1 | 50% | 0% | 1 | Search DOUBLE+CROSSOVERS | Search DOUBLE+CROSSOVERS |
| 10 | E COLI ADHESIN | 1 | 50% | 0% | 1 | Search E+COLI+ADHESIN | Search E+COLI+ADHESIN |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PERIODIC SELECTION | 9 | 52% | 3% | 12 |
| 2 | FUNDAMENTAL UNITS | 6 | 33% | 4% | 16 |
| 3 | BACTERIAL SPECIATION | 4 | 44% | 2% | 7 |
| 4 | MOLECULAR SEQUENCE | 3 | 100% | 1% | 3 |
| 5 | RANDOM DIVISION | 3 | 100% | 1% | 3 |
| 6 | GENETIC EXCHANGE | 3 | 11% | 5% | 22 |
| 7 | 2 MICROBIAL MATS | 2 | 50% | 1% | 3 |
| 8 | DNA SEQUENCE ALIGNMENTS | 2 | 40% | 1% | 4 |
| 9 | DETECTING RECOMBINATION | 2 | 12% | 4% | 16 |
| 10 | GRAZING SNAILS | 2 | 43% | 1% | 3 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| The Bacterial Species Challenge: Making Sense of Genetic and Ecological Diversity | 2009 | 151 | 31 | 55% |
| Horizontal gene transfer and the evolution of bacterial and archaeal population structure | 2013 | 47 | 46 | 41% |
| Recombination and the nature of bacterial speciation | 2007 | 193 | 28 | 57% |
| Explaining microbial genomic diversity in light of evolutionary ecology | 2014 | 19 | 97 | 24% |
| Microbial diversity and the genetic nature of microbial species | 2008 | 188 | 76 | 22% |
| Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches | 2011 | 100 | 145 | 19% |
| Ordering microbial diversity into ecologically and genetically cohesive units | 2014 | 7 | 73 | 44% |
| Bacterial species may exist, metagenomics reveal | 2012 | 26 | 39 | 41% |
| Re-evaluating prokaryotic species | 2005 | 442 | 37 | 22% |
| Impact of recombination on bacterial evolution | 2010 | 78 | 73 | 22% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROGRAM COMPUTAT SYST BIOL | 3 | 35% | 1.3% | 6 |
| 2 | BIOINFORMAT EVOLUTIONARY GENOM GRP | 1 | 33% | 0.4% | 2 |
| 3 | COMPUTAT GLOBAL SECUR | 1 | 50% | 0.2% | 1 |
| 4 | INFECT DIS VECTORS ECOL GENET EVOLUT CONTROL | 1 | 50% | 0.2% | 1 |
| 5 | UR0407 PATHOL VEGETALE | 1 | 50% | 0.2% | 1 |
| 6 | EVOLUC MOL EXPT | 1 | 19% | 0.7% | 3 |
| 7 | PARSONS ENVIRONM SCI ENGN | 1 | 22% | 0.4% | 2 |
| 8 | ROBERT CEDERGREN BIOCHIM | 0 | 33% | 0.2% | 1 |
| 9 | BIOHUMANITIES PROJECT | 0 | 25% | 0.2% | 1 |
| 10 | ENVIRONM INFECT RISKS UNIT | 0 | 25% | 0.2% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000175394 | TREE OF LIFE//HORIZONTAL GENE TRANSFER//LATERAL GENE TRANSFER |
| 2 | 0.0000164966 | KAUFFMANN WHITE SCHEME//I CEUI//HMU UCFM INFECT GENOM |
| 3 | 0.0000128637 | EVOLUTION CANYON//NALASSUS//BIODIVERSITY CHARACTERISTICS |
| 4 | 0.0000126803 | DISSOLVED DNA//NATURAL TRANSFORMATION//DNA PERSISTENCE |
| 5 | 0.0000096944 | PROTEIN SIP//SOIL METAPROTEOMICS//CHEM DIRECTORATE |
| 6 | 0.0000083677 | METAGENOMICS//MICROBIAL GENOM METAGEN PROGRAM//BIOSCI RD |
| 7 | 0.0000079033 | PHYLOGENETIC NETWORK//PHYLOGENETIC NETWORKS//TIGHT SPAN |
| 8 | 0.0000074949 | CODON MODEL//HETEROTACHY//PHYLOGENETIC INVARIANTS |
| 9 | 0.0000068741 | EXPERIMENTAL EVOLUTION//CLONAL INTERFERENCE//BENEFICIAL MUTATIONS |
| 10 | 0.0000057990 | ADAPTIVE MUTATION//STATIONARY PHASE MUTATION//STATIONARY PHASE MUTAGENESIS |