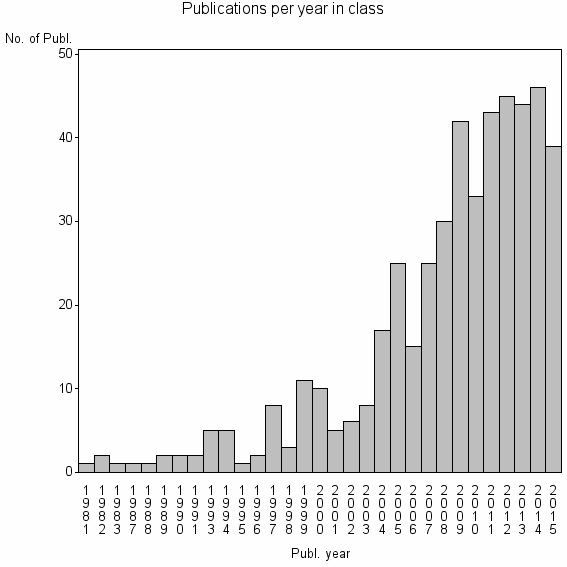
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 18572 | 480 | 37.2 | 77% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CORRELATED MUTATIONS | Author keyword | 11 | 47% | 4% | 17 |
| 2 | CONTACT PREDICTION | Author keyword | 7 | 53% | 2% | 10 |
| 3 | CONTACT MAP PREDICTION | Author keyword | 7 | 67% | 1% | 6 |
| 4 | INVERSE ISING PROBLEM | Author keyword | 6 | 100% | 1% | 4 |
| 5 | RESIDUE RESIDUE CONTACT | Author keyword | 4 | 67% | 1% | 4 |
| 6 | AMINO ACID CONTACTS | Author keyword | 4 | 75% | 1% | 3 |
| 7 | DIRECT COUPLING ANALYSIS | Author keyword | 4 | 75% | 1% | 3 |
| 8 | RESIDUE CONTACT PREDICTION | Author keyword | 4 | 75% | 1% | 3 |
| 9 | PROTEIN CONTACT MAP | Author keyword | 3 | 57% | 1% | 4 |
| 10 | CORRELATED SUBSTITUTIONS | Author keyword | 3 | 100% | 1% | 3 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CORRELATED MUTATIONS | 74 | 46% | 25% | 120 |
| 2 | RESIDUE CONTACTS | 61 | 62% | 13% | 63 |
| 3 | CONTACT PREDICTION | 26 | 56% | 6% | 31 |
| 4 | MULTIPLE SEQUENCE ALIGNMENTS | 13 | 20% | 12% | 59 |
| 5 | CORRELATED MUTATION ANALYSIS | 12 | 86% | 1% | 6 |
| 6 | ALLOSTERIC COMMUNICATION | 11 | 38% | 5% | 23 |
| 7 | COEVOLVING RESIDUES | 11 | 100% | 1% | 6 |
| 8 | COEVOLVING POSITIONS | 10 | 50% | 3% | 14 |
| 9 | INTERRESIDUE CONTACTS | 9 | 83% | 1% | 5 |
| 10 | BOLTZMANN MACHINES | 9 | 30% | 5% | 24 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Emerging methods in protein co-evolution | 2013 | 71 | 102 | 36% |
| Protein structure prediction from sequence variation | 2012 | 59 | 49 | 47% |
| Soft computing methods for the prediction of protein tertiary structures: A survey | 2015 | 0 | 76 | 49% |
| Evolution-Based Design of Proteins | 2013 | 6 | 25 | 28% |
| Protein Engineering and Stabilization from Sequence Statistics: Variation and Covariation Analysis | 2013 | 3 | 36 | 36% |
| Context dependence and coevolution among amino acid residues in proteins | 2005 | 16 | 29 | 48% |
| Inferring Pairwise Interactions from Biological Data Using Maximum-Entropy Probability Models | 2015 | 0 | 49 | 47% |
| Using Analyses of Amino Acid Coevolution to Understand Protein Structure and Function | 2013 | 1 | 55 | 40% |
| The functional importance of co-evolving residues in proteins | 2014 | 2 | 82 | 17% |
| Protein co-evolution: how do we combine bioinformatics and experimental approaches? | 2013 | 7 | 59 | 19% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EVOLUTIONARY GENET BIOINFORMAT | 2 | 33% | 0.8% | 4 |
| 2 | GENOM MICROORGANISMES | 1 | 20% | 1.3% | 6 |
| 3 | FINE CHEM ADV SYNTH CATALYSIS | 1 | 50% | 0.2% | 1 |
| 4 | GENETEVOLUTIONARY GENET BIOINFORMAT | 1 | 50% | 0.2% | 1 |
| 5 | GENETSMURFIT GENET | 1 | 50% | 0.2% | 1 |
| 6 | LECCE SPACI CONSORTIUM | 1 | 50% | 0.2% | 1 |
| 7 | MFG TEST | 1 | 50% | 0.2% | 1 |
| 8 | OURCE BIOCOMP VISUALIZAT INFORMAT | 1 | 50% | 0.2% | 1 |
| 9 | RWCP SNN | 1 | 50% | 0.2% | 1 |
| 10 | UMR CNRS INSERM 1083 6214 | 1 | 50% | 0.2% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000142901 | PROTEIN STRUCTURE PREDICTION//FOLD RECOGNITION//CASP |
| 2 | 0.0000126526 | SECONDARY STRUCTURE PREDICTION//PROTEIN SECONDARY STRUCTURE PREDICTION//COMPOUND PYRAMID MODEL |
| 3 | 0.0000120209 | HP MODEL//STATISTICAL POTENTIALS//PROTEIN STRUCTURE PREDICTION |
| 4 | 0.0000116514 | BLAST SEARCH//GENES FOR BASIC HELIX LOOP HELIX PROTEINS//ORTHOLOGOUS FAMILY |
| 5 | 0.0000103696 | MEAN FIELD VARIATIONAL BAYES//EXPECTATION PROPAGATION//GAUSSIAN PROCESSES MODEL |
| 6 | 0.0000089811 | ELASTIC NETWORK MODEL//ELASTIC NETWORK MODELS//NORMAL MODE ANALYSIS |
| 7 | 0.0000089550 | COMPLEX SCI SOFTWARE//3D ZERNIKE DESCRIPTOR//PROTEIN SURFACE SHAPE |
| 8 | 0.0000074492 | SATISFIABILITY//MESSAGE PASSING ALGORITHMS//CAVITY AND REPLICA METHOD |
| 9 | 0.0000060238 | DEEP LEARNING//HIERARCHICAL FEATURE LEARNING//RESTRICTED BOLTZMANN MACHINES |
| 10 | 0.0000058669 | CODON MODEL//HETEROTACHY//PHYLOGENETIC INVARIANTS |