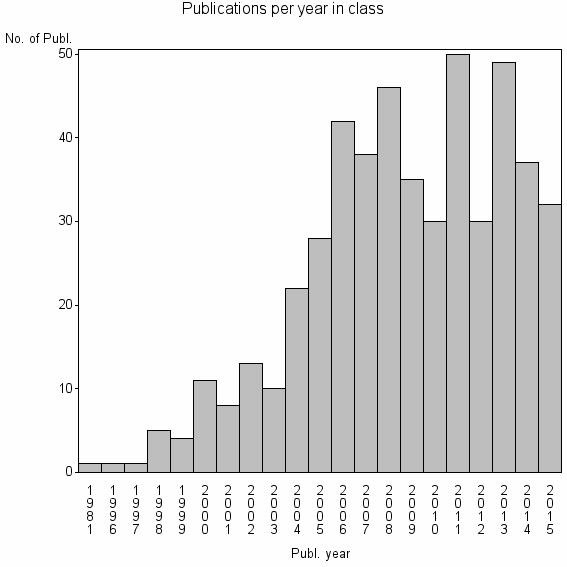
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 18304 | 493 | 30.5 | 83% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 238 | 19563 | BIOINFORMATICS//BMC BIOINFORMATICS//GENOME RESEARCH |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CIRCOS | Author keyword | 4 | 75% | 1% | 3 |
| 2 | BIODADOS | Address | 3 | 43% | 1% | 6 |
| 3 | ORTHOLOGY | Author keyword | 3 | 15% | 3% | 17 |
| 4 | BERKELEY PHYLOGENOM GRP | Address | 2 | 67% | 0% | 2 |
| 5 | BIDIRECTIONAL BEST HIT | Author keyword | 2 | 67% | 0% | 2 |
| 6 | GENOMIC DATA VISUALIZATION | Author keyword | 1 | 100% | 0% | 2 |
| 7 | RECIPROCAL BEST HIT | Author keyword | 1 | 100% | 0% | 2 |
| 8 | XENOLOG | Author keyword | 1 | 100% | 0% | 2 |
| 9 | BIOINFORMAT GENOM PROGRAMME | Address | 1 | 11% | 2% | 10 |
| 10 | DATA INTEGRAT ANAL IL | Address | 1 | 30% | 1% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CIRCOS | 4 | 75% | 1% | 3 | Search CIRCOS | Search CIRCOS |
| 2 | ORTHOLOGY | 3 | 15% | 3% | 17 | Search ORTHOLOGY | Search ORTHOLOGY |
| 3 | BIDIRECTIONAL BEST HIT | 2 | 67% | 0% | 2 | Search BIDIRECTIONAL+BEST+HIT | Search BIDIRECTIONAL+BEST+HIT |
| 4 | GENOMIC DATA VISUALIZATION | 1 | 100% | 0% | 2 | Search GENOMIC+DATA+VISUALIZATION | Search GENOMIC+DATA+VISUALIZATION |
| 5 | RECIPROCAL BEST HIT | 1 | 100% | 0% | 2 | Search RECIPROCAL+BEST+HIT | Search RECIPROCAL+BEST+HIT |
| 6 | XENOLOG | 1 | 100% | 0% | 2 | Search XENOLOG | Search XENOLOG |
| 7 | ORTHOLOG | 1 | 10% | 2% | 10 | Search ORTHOLOG | Search ORTHOLOG |
| 8 | ALL AGAINST ALL | 1 | 50% | 0% | 1 | Search ALL+AGAINST+ALL | Search ALL+AGAINST+ALL |
| 9 | ANNOTATION PIPELINES | 1 | 50% | 0% | 1 | Search ANNOTATION+PIPELINES | Search ANNOTATION+PIPELINES |
| 10 | AP1PMA | 1 | 50% | 0% | 1 | Search AP1PMA | Search AP1PMA |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PARALOGS | 12 | 35% | 6% | 29 |
| 2 | ORTHOMCL | 8 | 52% | 2% | 11 |
| 3 | ORTHOLOGY | 5 | 40% | 2% | 10 |
| 4 | COG DATABASE | 5 | 17% | 6% | 28 |
| 5 | ORTHOLOGS | 4 | 15% | 5% | 24 |
| 6 | AUTOMATED CONSTRUCTION | 3 | 57% | 1% | 4 |
| 7 | MULTIPLE GENOMES | 3 | 100% | 1% | 3 |
| 8 | BIOINFORMATICS RESOURCE UPDATE | 2 | 67% | 0% | 2 |
| 9 | CGVIEW | 2 | 67% | 0% | 2 |
| 10 | MBGD | 2 | 67% | 0% | 2 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Large-scale assignment of orthology: back to phylogenetics? | 2008 | 40 | 33 | 52% |
| Orthologs, paralogs, and evolutionary genomics | 2005 | 377 | 67 | 13% |
| In Silico Bioinformatic Tools for Determining Core Genes From Sets of Genomes | 2011 | 0 | 23 | 52% |
| The genome browser at UCSC for locating genes, and much more! | 2008 | 8 | 9 | 33% |
| Computer-Assisted Automatic Classifications, Storage, Queries and Functional Assignments of Orthologs and In-Paralogs Proteins | 2009 | 1 | 131 | 36% |
| Orthology and functional conservation in eukaryotes | 2007 | 26 | 53 | 25% |
| A reference guide for tree analysis and visualization | 2010 | 8 | 53 | 23% |
| Space, time, form: viewing the Tree of Life | 2012 | 5 | 36 | 19% |
| A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes | 2004 | 181 | 113 | 5% |
| Homology and phylogeny and their automated inference | 2008 | 2 | 72 | 21% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIODADOS | 3 | 43% | 1.2% | 6 |
| 2 | BERKELEY PHYLOGENOM GRP | 2 | 67% | 0.4% | 2 |
| 3 | BIOINFORMAT GENOM PROGRAMME | 1 | 11% | 2.0% | 10 |
| 4 | DATA INTEGRAT ANAL IL | 1 | 30% | 0.6% | 3 |
| 5 | CUBE COMPUTAT SYST BIOL | 1 | 33% | 0.4% | 2 |
| 6 | DWT | 1 | 50% | 0.2% | 1 |
| 7 | HEIDELBERG PERSONALIZED ONCOL DKFZ HIPO | 1 | 50% | 0.2% | 1 |
| 8 | HIPERSOFT | 1 | 50% | 0.2% | 1 |
| 9 | INRA BIOTECHNOL CHAMPIGNONS FILAMENTEUXUMR 1163 | 1 | 50% | 0.2% | 1 |
| 10 | LECCE SPACI CONSORTIUM | 1 | 50% | 0.2% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000195316 | STANDARDS IN GENOMIC SCIENCES//BIOL DATA MANAGEMENT TECHNOL//GEBA |
| 2 | 0.0000170142 | JOURNAL OF BIOMEDICAL SEMANTICS//ZFIN//ZEBRAFISH INFORMAT NETWORK |
| 3 | 0.0000157358 | EVOLUTIONARY BIOINFORMAT//CLOSED LOOPS//SKEW PAIRS |
| 4 | 0.0000153292 | WHOLE GENOME DUPLICATION//GENET MOL CELLULAIRE ECT LEVU INTERE//GENOME DUPLICATION |
| 5 | 0.0000150811 | EMBL OUTSTN//PROT INFORMAT OURCE//HLTH SCI TECHNOL ICIR |
| 6 | 0.0000143860 | TREE OF LIFE//HORIZONTAL GENE TRANSFER//LATERAL GENE TRANSFER |
| 7 | 0.0000137618 | PROGRAM COMPUTAT GENOM//CIFN//PROGRAMA GENOM COMPUTAC |
| 8 | 0.0000135503 | GENOME REARRANGEMENTS//SORTING BY REVERSALS//BREAKPOINT GRAPH |
| 9 | 0.0000112038 | PHYLOGENETIC NETWORK//PHYLOGENETIC NETWORKS//TIGHT SPAN |
| 10 | 0.0000103878 | MULTIPLE SEQUENCE ALIGNMENT//T COFFEE//STATISTICAL ALIGNMENT |