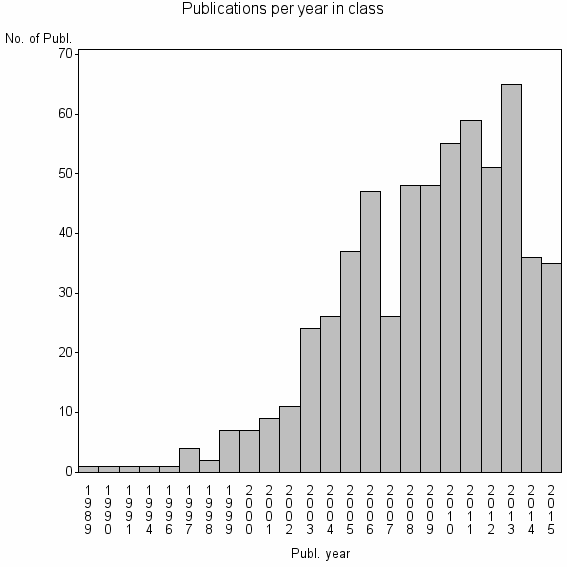
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 16178 | 602 | 35.9 | 67% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 584 | 13517 | BMC SYSTEMS BIOLOGY//FLUX BALANCE ANALYSIS//SYNTHETIC BIOLOGY |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | CELLML | Author keyword | 10 | 50% | 2% | 15 |
| 2 | BIOINFORMAT TUEBINGEN ZBIT | Address | 7 | 46% | 2% | 12 |
| 3 | SCI DATABASES VISUALIZAT GRP | Address | 6 | 80% | 1% | 4 |
| 4 | PHYSIOME PROJECT | Author keyword | 6 | 53% | 1% | 8 |
| 5 | COMPUTATIONAL PHYSIOLOGY | Author keyword | 5 | 41% | 1% | 9 |
| 6 | SBML | Author keyword | 4 | 33% | 2% | 11 |
| 7 | BIOMIMET MAT ENGN BIOL TISSUES | Address | 3 | 100% | 0% | 3 |
| 8 | FIELDML | Author keyword | 3 | 100% | 0% | 3 |
| 9 | MCT SET | Author keyword | 3 | 100% | 0% | 3 |
| 10 | MOLECULAR INTERACTION MAPS | Author keyword | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | CELLML | 10 | 50% | 2% | 15 | Search CELLML | Search CELLML |
| 2 | PHYSIOME PROJECT | 6 | 53% | 1% | 8 | Search PHYSIOME+PROJECT | Search PHYSIOME+PROJECT |
| 3 | COMPUTATIONAL PHYSIOLOGY | 5 | 41% | 1% | 9 | Search COMPUTATIONAL+PHYSIOLOGY | Search COMPUTATIONAL+PHYSIOLOGY |
| 4 | SBML | 4 | 33% | 2% | 11 | Search SBML | Search SBML |
| 5 | FIELDML | 3 | 100% | 0% | 3 | Search FIELDML | Search FIELDML |
| 6 | MCT SET | 3 | 100% | 0% | 3 | Search MCT+SET | Search MCT+SET |
| 7 | MOLECULAR INTERACTION MAPS | 3 | 100% | 0% | 3 | Search MOLECULAR+INTERACTION+MAPS | Search MOLECULAR+INTERACTION+MAPS |
| 8 | PHYSIOME | 3 | 24% | 2% | 10 | Search PHYSIOME | Search PHYSIOME |
| 9 | CARDIAC CELLULAR ELECTROPHYSIOLOGY | 2 | 67% | 0% | 2 | Search CARDIAC+CELLULAR+ELECTROPHYSIOLOGY | Search CARDIAC+CELLULAR+ELECTROPHYSIOLOGY |
| 10 | ONCO PROTEIN INHIBITORS | 2 | 67% | 0% | 2 | Search ONCO+PROTEIN+INHIBITORS | Search ONCO+PROTEIN+INHIBITORS |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SBML | 37 | 46% | 10% | 60 |
| 2 | PROTEIN INTERACTION DOMAINS | 9 | 83% | 1% | 5 |
| 3 | COMBINATORIAL COMPLEXITY | 7 | 35% | 3% | 17 |
| 4 | SOFTWARE ENVIRONMENT | 7 | 27% | 4% | 22 |
| 5 | MIRIAM | 6 | 80% | 1% | 4 |
| 6 | SBGN MAPS | 6 | 100% | 1% | 4 |
| 7 | QUANTITATIVE KINETIC MODELS | 5 | 60% | 1% | 6 |
| 8 | MARKUP LANGUAGE | 5 | 22% | 3% | 21 |
| 9 | CELLML | 5 | 43% | 1% | 9 |
| 10 | BIOLOGY MARKUP LANGUAGE | 5 | 37% | 2% | 11 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Software for systems biology: from tools to integrated platforms | 2011 | 62 | 84 | 20% |
| Computational systems biology in drug discovery and development: methods and applications | 2007 | 53 | 47 | 26% |
| Recent advances in modeling languages for pathway maps and computable biological networks | 2014 | 2 | 26 | 27% |
| Standards and ontologies in computational systems biology | 2008 | 8 | 18 | 78% |
| Model storage, exchange and integration | 2006 | 28 | 9 | 67% |
| Minimum Information about a Cardiac Electrophysiology Experiment (MICEE): Standardised reporting for model reproducibility, interoperability, and data sharing | 2011 | 16 | 21 | 33% |
| The Virtual Cell: a software environment for computational cell biology | 2001 | 154 | 15 | 27% |
| Connecting the dots: role of standardization and technology sharing in biological simulation | 2010 | 1 | 9 | 78% |
| A review of standards for data exchange within systems biology | 2007 | 22 | 18 | 33% |
| OpenCMISS: A multi-physics & multi-scale computational infrastructure for the VPH/Physiome project | 2011 | 20 | 37 | 24% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BIOINFORMAT TUEBINGEN ZBIT | 7 | 46% | 2.0% | 12 |
| 2 | SCI DATABASES VISUALIZAT GRP | 6 | 80% | 0.7% | 4 |
| 3 | BIOMIMET MAT ENGN BIOL TISSUES | 3 | 100% | 0.5% | 3 |
| 4 | FISIOCOMP | 2 | 67% | 0.3% | 2 |
| 5 | GERMAN VIRTUAL LIVER NETWORK | 2 | 67% | 0.3% | 2 |
| 6 | GRP CLIN GENOM NETWORKS | 2 | 67% | 0.3% | 2 |
| 7 | BIO GEOSCI BIOTECHNOL 1 | 1 | 50% | 0.3% | 2 |
| 8 | MAASTRICHT SYST BIOL MACSBIO | 1 | 50% | 0.3% | 2 |
| 9 | CELL ANAL MODELING | 1 | 10% | 2.0% | 12 |
| 10 | BIOL NETWORK MODELING | 1 | 30% | 0.5% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000189076 | FLUX BALANCE ANALYSIS//CONSTRAINT BASED MODELING//METABOLIC NETWORK |
| 2 | 0.0000170382 | BIOCHEMICAL SYSTEMS THEORY//S SYSTEM//S DISTRIBUTION |
| 3 | 0.0000158448 | MASS ACTION KINETICS//COMPLEX BALANCING//CHEMICAL REACTION NETWORKS |
| 4 | 0.0000115561 | CELL DECIS PROC//MATH ARTS SCI//SIGNALING SYST |
| 5 | 0.0000114896 | JOURNAL OF BIOMEDICAL SEMANTICS//ZFIN//ZEBRAFISH INFORMAT NETWORK |
| 6 | 0.0000108073 | BOOLEAN NETWORKS//SEMI TENSOR PRODUCT//BOOLEAN NETWORK |
| 7 | 0.0000098552 | EUFORIA//DISTRIBUTED MULTISCALE COMPUTING//COMPUTAT LIFE MED SCI NETWORK |
| 8 | 0.0000089548 | LINEAR NOISE APPROXIMATION//STOCHASTIC GENE EXPRESSION//CHEMICAL MASTER EQUATION |
| 9 | 0.0000089461 | METABOLIC CONTROL ANALYSIS//CONTROL COEFFICIENT//CONTROL COEFFICIENTS |
| 10 | 0.0000074840 | ROBERT ROSEN//M R SYSTEMS//RELATIONAL BIOLOGY |