Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
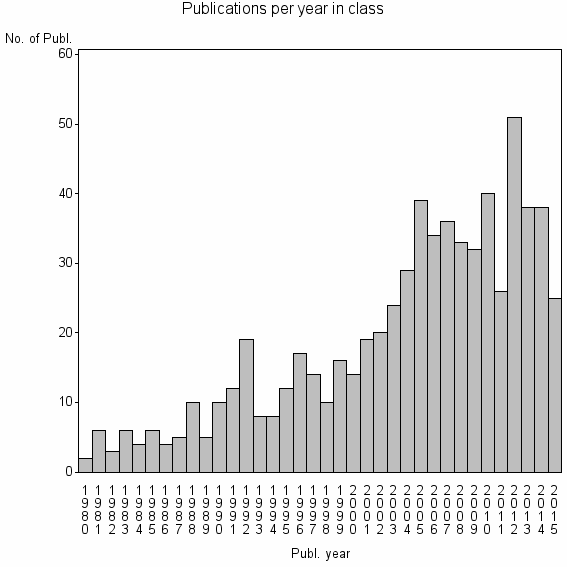
| 14877 | 675 | 45.6 | 82% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1971 | 5134 | O ANTIGEN//BACTERIAL POLYSACCHARIDE STRUCTURE//O SPECIFIC POLYSACCHARIDE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | LPXC | Author keyword | 18 | 65% | 3% | 17 |
| 2 | LPXC INHIBITORS | Author keyword | 15 | 88% | 1% | 7 |
| 3 | LPXA | Author keyword | 8 | 70% | 1% | 7 |
| 4 | LIPID A BIOSYNTHESIS | Author keyword | 7 | 67% | 1% | 6 |
| 5 | MGTA | Author keyword | 6 | 80% | 1% | 4 |
| 6 | MIDDLE ATLANTIC MASS SPE OMETRY | Address | 6 | 38% | 2% | 12 |
| 7 | PHOP PHOQ | Author keyword | 6 | 50% | 1% | 8 |
| 8 | DUKE NMR SPECT | Address | 5 | 45% | 1% | 9 |
| 9 | PMRD | Author keyword | 4 | 67% | 1% | 4 |
| 10 | HLTH SCI 4H19 | Address | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | LPXC | 18 | 65% | 3% | 17 | Search LPXC | Search LPXC |
| 2 | LPXC INHIBITORS | 15 | 88% | 1% | 7 | Search LPXC+INHIBITORS | Search LPXC+INHIBITORS |
| 3 | LPXA | 8 | 70% | 1% | 7 | Search LPXA | Search LPXA |
| 4 | LIPID A BIOSYNTHESIS | 7 | 67% | 1% | 6 | Search LIPID+A+BIOSYNTHESIS | Search LIPID+A+BIOSYNTHESIS |
| 5 | MGTA | 6 | 80% | 1% | 4 | Search MGTA | Search MGTA |
| 6 | PHOP PHOQ | 6 | 50% | 1% | 8 | Search PHOP+PHOQ | Search PHOP+PHOQ |
| 7 | PMRD | 4 | 67% | 1% | 4 | Search PMRD | Search PMRD |
| 8 | PMRA PMRB | 3 | 100% | 0% | 3 | Search PMRA+PMRB | Search PMRA+PMRB |
| 9 | LPXL | 3 | 60% | 0% | 3 | Search LPXL | Search LPXL |
| 10 | MGTC | 3 | 60% | 0% | 3 | Search MGTC | Search MGTC |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | ENDOTOXIN BIOSYNTHESIS | 96 | 86% | 7% | 49 |
| 2 | ZINC DEPENDENT DEACETYLASE | 70 | 86% | 5% | 36 |
| 3 | UDP 3 O R 3 HYDROXYMYRISTOYL N ACETYLGLUCOSAMINE DEACETYLASE | 55 | 87% | 4% | 27 |
| 4 | POLYMYXIN RESISTANCE | 42 | 53% | 8% | 56 |
| 5 | LIPID A BIOSYNTHESIS | 31 | 57% | 5% | 37 |
| 6 | PHOP PHOQ | 30 | 37% | 10% | 66 |
| 7 | ANTIMICROBIAL PEPTIDE RESISTANCE | 26 | 49% | 6% | 38 |
| 8 | ACID DEFICIENT MUTANT | 21 | 73% | 2% | 16 |
| 9 | TEMPERATURE REQUIREMENT GENE | 17 | 68% | 2% | 15 |
| 10 | 3 O DEACYLATION | 17 | 100% | 1% | 8 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Biosynthesis and Export of Bacterial Lipopolysaccharides | 2014 | 17 | 204 | 50% |
| Lipid a modification systems in gram-negative bacteria | 2007 | 329 | 237 | 50% |
| Lipopolysaccharide: Biosynthetic pathway and structure modification | 2010 | 77 | 117 | 55% |
| Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis | 2013 | 30 | 143 | 39% |
| The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more | 2008 | 90 | 64 | 63% |
| Diversity of endotoxin and its impact on pathogenesis | 2006 | 119 | 178 | 49% |
| Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity | 2014 | 5 | 85 | 44% |
| The Biology of the PmrA/PmrB Two-Component System: The Major Regulator of Lipopolysaccharide Modifications | 2013 | 11 | 164 | 50% |
| Mechanism and inhibition of LpxC: An essential zinc-dependent deacetylase of bacterial lipid a synthesis | 2008 | 31 | 35 | 74% |
| UDP-3-O-(R-3-hydroxymyristoyl)-N-Acetylglucosamine Deacetylase (LpxC) Inhibitors: A New Class of Antibacterial Agents | 2012 | 10 | 55 | 62% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MIDDLE ATLANTIC MASS SPE OMETRY | 6 | 38% | 1.8% | 12 |
| 2 | DUKE NMR SPECT | 5 | 45% | 1.3% | 9 |
| 3 | HLTH SCI 4H19 | 3 | 100% | 0.4% | 3 |
| 4 | MIDDLE ATLANTIC MASS SPECTOMETRY | 1 | 100% | 0.3% | 2 |
| 5 | MIDDLE ATLANTIC MASS SPECT | 1 | 40% | 0.3% | 2 |
| 6 | STRUCT BIOL BIOPHYS PROGRAM | 1 | 17% | 0.6% | 4 |
| 7 | ANTIBACTERIALS UNIT | 1 | 21% | 0.4% | 3 |
| 8 | ANIM INFECT DIS MINIST EDUC | 1 | 50% | 0.1% | 1 |
| 9 | DISCOVERY TECHNOL STRUCT ANALYT SCI | 1 | 50% | 0.1% | 1 |
| 10 | DUKE NMR | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000237768 | KDO8P SYNTHASE//3 DEOXY D ARABINO HEPTULOSONATE 7 PHOSPHATE SYNTHASE//KDO8PS |
| 2 | 0.0000218586 | LIPID A ANALOG//LIPID A//LPS ANTAGONIST |
| 3 | 0.0000140842 | COLISTIN//POLYMYXINS//COLISTIMETHATE SODIUM |
| 4 | 0.0000123893 | SALMONELLA CONTAINING VACUOLE//SOPB//D BIOL |
| 5 | 0.0000113270 | AUTOTRANSPORTER//AUTODISPLAY//BAM COMPLEX |
| 6 | 0.0000091280 | O ANTIGEN//BACTERIAL POLYSACCHARIDE STRUCTURE//O ANTIGEN GENE CLUSTER |
| 7 | 0.0000087702 | ENVZ//OMPR//ENVZ OSMOSENSOR |
| 8 | 0.0000076339 | GLMU//UDP GLUCOSE PYROPHOSPHORYLASE//UDP SUGAR PYROPHOSPHORYLASE |
| 9 | 0.0000071892 | MD 2//CD14//LIPOPOLYSACCHARIDE BINDING PROTEIN |
| 10 | 0.0000060961 | ENOYL ACP REDUCTASE//ACYL CARRIER PROTEIN//FABH |