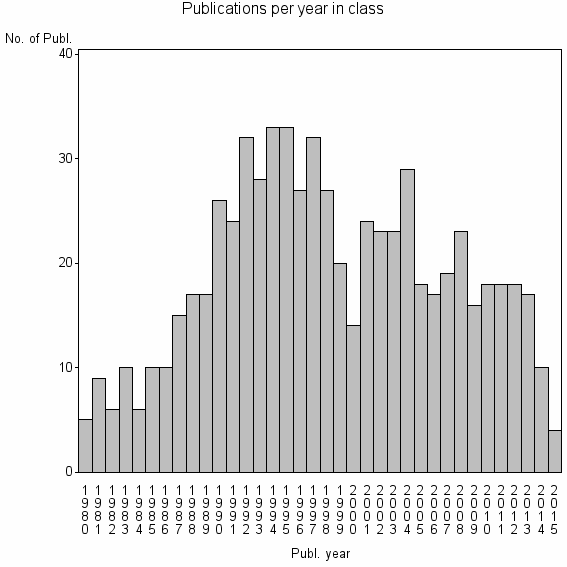
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 14837 | 678 | 40.3 | 72% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 1431 | 7403 | PHENYLKETONURIA//TETRAHYDROBIOPTERIN//HYPERPHENYLALANINEMIA |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PHENYLETHANOLAMINE N METHYLTRANSFERASE | Author keyword | 19 | 35% | 7% | 45 |
| 2 | PSYCHIAT MOL DEV NEUROBIOL | Address | 12 | 86% | 1% | 6 |
| 3 | TYROSINE HYDROXYLASE GENE | Author keyword | 7 | 48% | 2% | 11 |
| 4 | PNMT | Author keyword | 7 | 32% | 3% | 18 |
| 5 | CATECHOLAMINE BIOSYNTHETIC ENZYMES | Author keyword | 7 | 67% | 1% | 6 |
| 6 | NOVEL STRESSORS | Author keyword | 6 | 80% | 1% | 4 |
| 7 | ADRENERGIC EXPRESSION | Author keyword | 6 | 100% | 1% | 4 |
| 8 | DOPAMINE BETA HYDROXYLASE | Author keyword | 5 | 11% | 7% | 47 |
| 9 | PHENYLETHANOLAMINE N METHYLTRANSFERASE PNMT | Author keyword | 4 | 50% | 1% | 6 |
| 10 | MOL DEV NEUROBIOL | Address | 4 | 46% | 1% | 6 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PHENYLETHANOLAMINE N METHYLTRANSFERASE | 24 | 25% | 12% | 84 |
| 2 | CATECHOLAMINE BIOSYNTHETIC ENZYMES | 18 | 46% | 4% | 29 |
| 3 | REPEATED IMMOBILIZATION STRESS | 14 | 35% | 5% | 33 |
| 4 | PNMT MESSENGER RNA | 12 | 86% | 1% | 6 |
| 5 | TRANS SYNAPTIC INDUCTION | 8 | 100% | 1% | 5 |
| 6 | PHEOCHROMOCYTOMA CELL LINE | 7 | 40% | 2% | 14 |
| 7 | PNMT GENE | 6 | 71% | 1% | 5 |
| 8 | PARALLEL UP REGULATION | 6 | 53% | 1% | 8 |
| 9 | GENE TRANSCRIPTION RATE | 6 | 100% | 1% | 4 |
| 10 | RARE ALLELE | 6 | 100% | 1% | 4 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Catecholaminergic Systems in Stress: Structural and Molecular Genetic Approaches | 2009 | 140 | 563 | 27% |
| Stress-triggered activation of gene expression in catecholaminergic systems: dynamics of transcriptional events | 2001 | 151 | 69 | 64% |
| Epigenetic, transcriptional and posttranscriptional regulation of the tyrosine hydroxylase gene | 2011 | 15 | 159 | 50% |
| Epinephrine: A Short- and Long-Term Regulator of Stress and Development of Illness A Potential New Role for Epinephrine in Stress | 2012 | 17 | 58 | 36% |
| Control of tyrosine hydroxylase gene expression in chromaffin and PC12 cells | 1997 | 30 | 62 | 73% |
| Multiple signalling pathways exist in the stress-triggered regulation of gene expression for catecholamine biosynthetic enzymes and several neuropeptides in the rat adrenal medulla | 1999 | 31 | 55 | 55% |
| Catecholamine biosynthesis and physiological regulation in neuroendocrine cells | 2000 | 62 | 90 | 30% |
| Why is the adrenal adrenergic? | 2003 | 19 | 60 | 45% |
| Sympathoadrenal system in stress - Interaction with the hypothalamic-pituitary-adrenocortical system | 1995 | 137 | 79 | 28% |
| Activity-dependent regulation of the dopamine phenotype in substantia nigra neurons | 2012 | 7 | 181 | 15% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PSYCHIAT MOL DEV NEUROBIOL | 12 | 86% | 0.9% | 6 |
| 2 | MOL DEV NEUROBIOL | 4 | 46% | 0.9% | 6 |
| 3 | CHGG | 2 | 67% | 0.3% | 2 |
| 4 | MOL DEV NEUROBIOLMCLEAN HOSP | 1 | 100% | 0.3% | 2 |
| 5 | REG NEONATAL | 1 | 17% | 0.9% | 6 |
| 6 | GENET NEUROTRANSMISS | 1 | 33% | 0.3% | 2 |
| 7 | UMR C9923 | 1 | 21% | 0.4% | 3 |
| 8 | ANESTHESIOLCHILDRENS HOSP BOSTON | 1 | 50% | 0.1% | 1 |
| 9 | EDUC CLIN 182 | 1 | 50% | 0.1% | 1 |
| 10 | EXPT ALCOHOL DRUG SECT | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000241829 | PHENYLALANINE HYDROXYLASE//TRYPTOPHAN HYDROXYLASE//AROMATIC AMINO ACID HYDROXYLASE |
| 2 | 0.0000149805 | 2 3 4 5 TETRAHYDRO 1H 2 AZEPINE//2 3 4 5 TETRAHYDRO 1H 3 BENZAZEPINE//A RING CATECHOL |
| 3 | 0.0000104343 | CONGENITAL CENTRAL HYPOVENTILATION SYNDROME//PHOX2B//PHOX2B GENE |
| 4 | 0.0000095753 | ORNITHINE DECARBOXYLASE IN DEVELOPING BRAIN//ADENYLYL CYCLASE SIGNALING//NORADRENERGIC SYSTEMS |
| 5 | 0.0000073408 | PROENKEPHALIN//PROENKEPHALINS//PRO ENKEPHALIN |
| 6 | 0.0000068666 | AROMATIC L AMINO ACID DECARBOXYLASE//LABORATORY SHREW//DOPA CYCLOHEXYL ESTER |
| 7 | 0.0000067033 | CILIARY NEUROTROPHIC FACTOR//CNTF//CNTFR ALPHA |
| 8 | 0.0000066479 | UNITAT FISIOL ANIM//UNITAT PSICOBIOL//NOISE STRESS |
| 9 | 0.0000064632 | CYTOCHROME B561//PEPTIDYLGLYCINE ALPHA AMIDATING MONOOXYGENASE//PEPTIDYLGLYCINE ALPHA HYDROXYLATING MONOOXYGENASE |
| 10 | 0.0000063451 | PUSH PULL SUPERFUSION//RU24722//PUSH PULL CANNULA |