Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
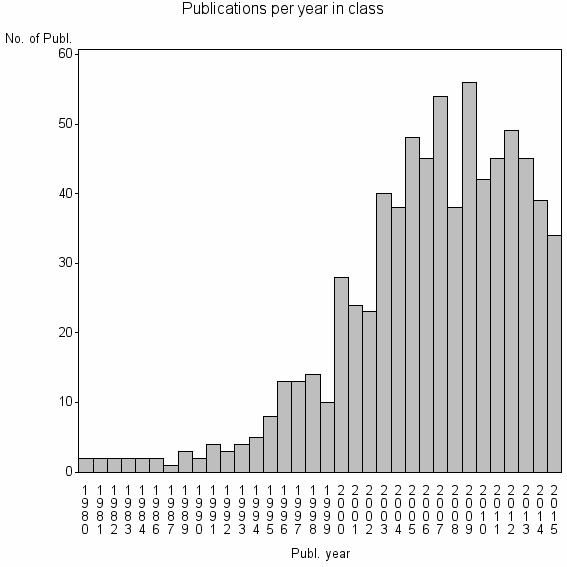
| 13926 | 740 | 40.9 | 82% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 77 | 26633 | NUCLEOSIDES NUCLEOTIDES & NUCLEIC ACIDS//NUCLEOSIDES & NUCLEOTIDES//NUCLEOSIDES |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | METAL MEDIATED BASE PAIRS | Author keyword | 26 | 100% | 1% | 11 |
| 2 | METAL MEDIATED BASE PAIR | Author keyword | 11 | 100% | 1% | 6 |
| 3 | UNNATURAL BASE PAIR | Author keyword | 11 | 100% | 1% | 6 |
| 4 | PROT NUCLE ACID | Address | 8 | 70% | 1% | 7 |
| 5 | HYDROPHOBIC BASES | Author keyword | 6 | 80% | 1% | 4 |
| 6 | GENETIC ALPHABET | Author keyword | 6 | 71% | 1% | 5 |
| 7 | UNNATURAL BASE PAIRS | Author keyword | 5 | 63% | 1% | 5 |
| 8 | SSBC | Address | 4 | 67% | 1% | 4 |
| 9 | MOL TRANSFORMAT | Address | 3 | 39% | 1% | 7 |
| 10 | METAL BASE PAIRS | Author keyword | 3 | 100% | 0% | 3 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | METAL MEDIATED BASE PAIRS | 26 | 100% | 1% | 11 | Search METAL+MEDIATED+BASE+PAIRS | Search METAL+MEDIATED+BASE+PAIRS |
| 2 | METAL MEDIATED BASE PAIR | 11 | 100% | 1% | 6 | Search METAL+MEDIATED+BASE+PAIR | Search METAL+MEDIATED+BASE+PAIR |
| 3 | UNNATURAL BASE PAIR | 11 | 100% | 1% | 6 | Search UNNATURAL+BASE+PAIR | Search UNNATURAL+BASE+PAIR |
| 4 | HYDROPHOBIC BASES | 6 | 80% | 1% | 4 | Search HYDROPHOBIC+BASES | Search HYDROPHOBIC+BASES |
| 5 | GENETIC ALPHABET | 6 | 71% | 1% | 5 | Search GENETIC+ALPHABET | Search GENETIC+ALPHABET |
| 6 | UNNATURAL BASE PAIRS | 5 | 63% | 1% | 5 | Search UNNATURAL+BASE+PAIRS | Search UNNATURAL+BASE+PAIRS |
| 7 | METAL BASE PAIRS | 3 | 100% | 0% | 3 | Search METAL+BASE+PAIRS | Search METAL+BASE+PAIRS |
| 8 | C NUCLEOSIDES | 3 | 17% | 2% | 15 | Search C+NUCLEOSIDES | Search C+NUCLEOSIDES |
| 9 | ARTIFICIAL DNA | 2 | 38% | 1% | 5 | Search ARTIFICIAL+DNA | Search ARTIFICIAL+DNA |
| 10 | NONPOLAR ISOSTERE | 2 | 67% | 0% | 2 | Search NONPOLAR+ISOSTERE | Search NONPOLAR+ISOSTERE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | HYDROPHOBIC BASE | 157 | 87% | 10% | 76 |
| 2 | GENETIC ALPHABET | 148 | 64% | 20% | 145 |
| 3 | POLYMERASE RECOGNITION | 75 | 83% | 6% | 43 |
| 4 | ARTIFICIAL DNA | 51 | 57% | 8% | 60 |
| 5 | PAIRED GENETIC HELIX | 47 | 88% | 3% | 22 |
| 6 | ENZYMATIC INCORPORATION | 44 | 37% | 13% | 95 |
| 7 | DIFLUOROTOLUENE | 30 | 64% | 4% | 29 |
| 8 | SIZE EXPANDED DNA | 26 | 80% | 2% | 16 |
| 9 | UNNATURAL BASE PAIRS | 23 | 59% | 4% | 26 |
| 10 | MODIFIED PNA DUPLEXES | 21 | 69% | 2% | 18 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Metal-Mediated DNA Base Pairing: Alternatives to Hydrogen-Bonded Watson-Crick Base Pairs | 2012 | 74 | 40 | 75% |
| Natural versus Artificial Creation of Base Pairs in DNA: Origin of Nucleobases from the Perspectives of Unnatural Base Pair Studies | 2012 | 29 | 57 | 79% |
| Binding of metal ions by pyrimidine base pairs in DNA duplexes | 2011 | 68 | 64 | 50% |
| Nucleic Acids With Metal-Mediated Base Pairs and Their Applications | 2013 | 22 | 102 | 52% |
| C-Nucleosides: Synthetic Strategies and Biological Applications | 2009 | 85 | 363 | 45% |
| Metal-base pairing in DNA | 2010 | 77 | 85 | 45% |
| Unnatural base pair systems for DNA/RNA-based biotechnology | 2006 | 76 | 29 | 97% |
| DNA-metal base pairs | 2007 | 273 | 94 | 27% |
| Alternative DNA base-pairs: from efforts to expand the genetic code to potential material applications | 2011 | 28 | 75 | 72% |
| Beyond A, C, G and T: augmenting nature's alphabet | 2003 | 177 | 40 | 65% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROT NUCLE ACID | 8 | 70% | 0.9% | 7 |
| 2 | SSBC | 4 | 67% | 0.5% | 4 |
| 3 | MOL TRANSFORMAT | 3 | 39% | 0.9% | 7 |
| 4 | MODELING SIMULAT CHEM | 2 | 24% | 1.2% | 9 |
| 5 | YOKOYAMA CYTOLOG PROJECT | 1 | 25% | 0.7% | 5 |
| 6 | COMPUTAC BIOL PROGRAM | 1 | 100% | 0.3% | 2 |
| 7 | WESTHEIMER SCI TECHNOL | 1 | 50% | 0.3% | 2 |
| 8 | GILEAD IOCB | 1 | 18% | 0.9% | 7 |
| 9 | UNITAT MODELITZACIO MOL BIOINFORMAT | 1 | 33% | 0.4% | 3 |
| 10 | CHIM PHYS SCI | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000216176 | ORGAN BIOORGAN CHEM//BIOORGAN CHEM CHEM BIOL//7 DEAZAPURINE |
| 2 | 0.0000186436 | FLEXIMERS//4 METHYL BENZENESUFONYL CHLORIDE//INDOLE NUCLEOSIDE |
| 3 | 0.0000173364 | NEWCAT//MOLECULAR BEACON//MOLECULAR BEACONS |
| 4 | 0.0000156934 | 2 3 DIKETO L GULONIC ACID//C NUCLEOSIDES//PYRAZOLO4 3 BPYRIDINE |
| 5 | 0.0000153794 | DNA POLYMERASE//DNA POLYMERASE BETA//DNA POLYMERASE LAMBDA |
| 6 | 0.0000128465 | GILEAD IOCB//REVERSIBLE TERMINATOR//ONE POT METHODOLOGY |
| 7 | 0.0000100841 | C GLYCOSIDATIONS//ESMODIL//GLUCOSE DIACETONIDE |
| 8 | 0.0000091718 | PROGRAM BIOCHEM CHEM//RNA CRYSTALLOGRAPHY//5 METHYL CYTIDINE |
| 9 | 0.0000087337 | DNA ELECTRON TRANSFER//DNA CHARGE TRANSPORT//SCI IND SANKEN |
| 10 | 0.0000086136 | COLORIMETRIC DETECTION//MERCURY ION//MERCURY IONS |