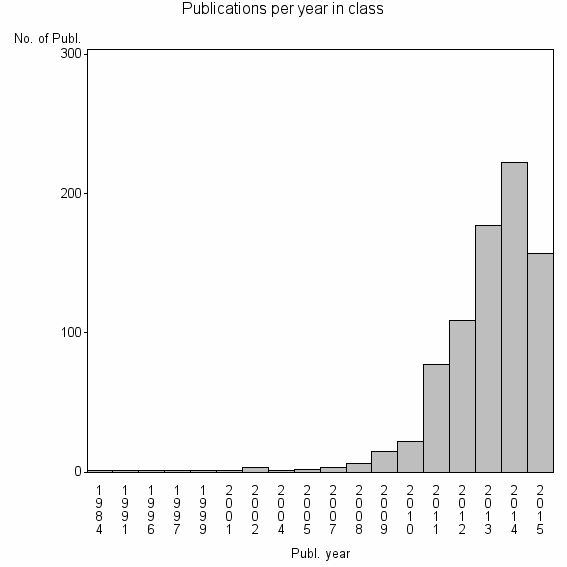
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 13046 | 800 | 54.1 | 94% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 348 | 16958 | DNA METHYLATION//EPIGENETICS//HISTONE DEACETYLASE INHIBITOR |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | 5 HYDROXYMETHYLCYTOSINE | Author keyword | 92 | 65% | 11% | 87 |
| 2 | 5HMC | Author keyword | 37 | 81% | 3% | 22 |
| 3 | 5 CARBOXYLCYTOSINE | Author keyword | 24 | 91% | 1% | 10 |
| 4 | TET PROTEINS | Author keyword | 23 | 79% | 2% | 15 |
| 5 | TET1 | Author keyword | 23 | 72% | 2% | 18 |
| 6 | 5 FORMYLCYTOSINE | Author keyword | 21 | 90% | 1% | 9 |
| 7 | 5 HYDROXYMETHYLCYTOSINE 5HMC | Author keyword | 21 | 85% | 1% | 11 |
| 8 | 5 METHYLCYTOSINE 5MC | Author keyword | 18 | 89% | 1% | 8 |
| 9 | HYDROXYMETHYLCYTOSINE | Author keyword | 17 | 75% | 2% | 12 |
| 10 | DNA DEMETHYLATION | Author keyword | 13 | 26% | 5% | 42 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | 5 HYDROXYMETHYLCYTOSINE | 92 | 65% | 11% | 87 | Search 5+HYDROXYMETHYLCYTOSINE | Search 5+HYDROXYMETHYLCYTOSINE |
| 2 | 5HMC | 37 | 81% | 3% | 22 | Search 5HMC | Search 5HMC |
| 3 | 5 CARBOXYLCYTOSINE | 24 | 91% | 1% | 10 | Search 5+CARBOXYLCYTOSINE | Search 5+CARBOXYLCYTOSINE |
| 4 | TET PROTEINS | 23 | 79% | 2% | 15 | Search TET+PROTEINS | Search TET+PROTEINS |
| 5 | TET1 | 23 | 72% | 2% | 18 | Search TET1 | Search TET1 |
| 6 | 5 FORMYLCYTOSINE | 21 | 90% | 1% | 9 | Search 5+FORMYLCYTOSINE | Search 5+FORMYLCYTOSINE |
| 7 | 5 HYDROXYMETHYLCYTOSINE 5HMC | 21 | 85% | 1% | 11 | Search 5+HYDROXYMETHYLCYTOSINE+5HMC | Search 5+HYDROXYMETHYLCYTOSINE+5HMC |
| 8 | 5 METHYLCYTOSINE 5MC | 18 | 89% | 1% | 8 | Search 5+METHYLCYTOSINE+5MC | Search 5+METHYLCYTOSINE+5MC |
| 9 | HYDROXYMETHYLCYTOSINE | 17 | 75% | 2% | 12 | Search HYDROXYMETHYLCYTOSINE | Search HYDROXYMETHYLCYTOSINE |
| 10 | DNA DEMETHYLATION | 13 | 26% | 5% | 42 | Search DNA+DEMETHYLATION | Search DNA+DEMETHYLATION |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | 5 HYDROXYMETHYLCYTOSINE | 257 | 58% | 37% | 294 |
| 2 | MAMMALIAN DNA | 223 | 60% | 30% | 242 |
| 3 | TET PROTEINS | 181 | 74% | 17% | 133 |
| 4 | 5 CARBOXYLCYTOSINE | 179 | 78% | 15% | 118 |
| 5 | 5 METHYLCYTOSINE | 134 | 35% | 39% | 308 |
| 6 | 5 FORMYLCYTOSINE | 124 | 85% | 8% | 66 |
| 7 | 5 METHYLCYTOSINE OXIDATION | 85 | 100% | 3% | 26 |
| 8 | BASE RESOLUTION | 79 | 74% | 7% | 59 |
| 9 | TET1 | 69 | 73% | 7% | 52 |
| 10 | THYMINE DNA | 67 | 78% | 6% | 45 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Reversing DNA Methylation: Mechanisms, Genomics, and Biological Functions | 2014 | 104 | 209 | 64% |
| Function and information content of DNA methylation | 2015 | 26 | 115 | 28% |
| TET enzymes, TDG and the dynamics of DNA demethylation | 2013 | 148 | 99 | 76% |
| Uncovering the role of 5-hydroxymethylcytosine in the epigenome | 2012 | 150 | 52 | 83% |
| Mechanism and Function of Oxidative Reversal of DNA and RNA Methylation | 2014 | 22 | 172 | 70% |
| TETonic shift: biological roles of TET proteins in DNA demethylation and transcription | 2013 | 113 | 179 | 59% |
| Playing TETris with DNA modifications | 2014 | 16 | 108 | 75% |
| The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases | 2014 | 25 | 108 | 53% |
| TET proteins and 5-methylcytosine oxidation in hematological cancers | 2015 | 5 | 159 | 46% |
| Chemical Methods for Decoding Cytosine Modifications in DNA | 2015 | 4 | 88 | 50% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | EPIGENET PROGRAMME | 5 | 27% | 2.0% | 16 |
| 2 | GRP DNA METAB | 4 | 75% | 0.4% | 3 |
| 3 | GRP DNA METABSTATE MOL BIOL | 3 | 100% | 0.4% | 3 |
| 4 | SIGNALING GENE EXP S | 3 | 33% | 0.9% | 7 |
| 5 | BIODYNAM OPT IMAGING | 2 | 17% | 1.5% | 12 |
| 6 | JOHN P HUSSMAN HUMAN GENOM HUMAN GENET | 2 | 67% | 0.3% | 2 |
| 7 | BIOSNS | 1 | 50% | 0.3% | 2 |
| 8 | EPIGENET MECH | 1 | 50% | 0.3% | 2 |
| 9 | HUMAN ENVIRONM EPIGENOMES | 1 | 100% | 0.3% | 2 |
| 10 | HUMAN REPROD DEV UNIT | 1 | 100% | 0.3% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000209433 | RRBS//CPG ISLANDS//METHYLTRANSFERASE ACTIVITY |
| 2 | 0.0000195417 | UHRF1//ICBP90//UHRF2 |
| 3 | 0.0000166785 | DNMT3B//DNA METHYLTRANSFERASE//DNMT1 |
| 4 | 0.0000134740 | ALKB//METTL3//ALKBH3 |
| 5 | 0.0000129414 | SACKLER PROGRAM EPIGENET DEV PSYCHOBIOL//AGING COGNIT DIS//BONNEY 106 |
| 6 | 0.0000083866 | TRANSGENERATIONAL//METASTABLE EPIALLELE//TRANSGENERATIONAL EPIGENETIC INHERITANCE |
| 7 | 0.0000083688 | IDH1//IDH2//ISOCITRATE DEHYDROGENASE 1 |
| 8 | 0.0000081607 | EPIGENETIC EPIDEMIOLOGY//EWAS//EPIGENETICS |
| 9 | 0.0000078735 | URACIL DNA GLYCOSYLASE//DUTPASE//DUTP PYROPHOSPHATASE |
| 10 | 0.0000063229 | VSELS//PRIMORDIAL GERM CELLS//VSEL |