Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
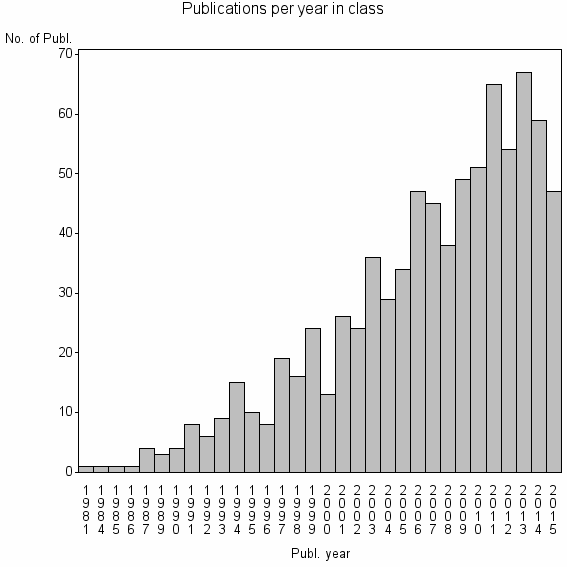
| 12864 | 814 | 45.4 | 86% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | COMPUTATIONAL PROTEIN DESIGN | Author keyword | 102 | 73% | 10% | 78 |
| 2 | DEAD END ELIMINATION | Author keyword | 40 | 76% | 3% | 28 |
| 3 | COMPUTATIONAL ENZYME DESIGN | Author keyword | 30 | 100% | 1% | 12 |
| 4 | PROTEIN DESIGN | Author keyword | 22 | 16% | 16% | 129 |
| 5 | SIDE CHAIN PREDICTION | Author keyword | 16 | 65% | 2% | 15 |
| 6 | ROTAMER LIBRARY | Author keyword | 16 | 58% | 2% | 18 |
| 7 | SIDE CHAIN MODELING | Author keyword | 11 | 69% | 1% | 9 |
| 8 | FLEXIBLE BACKBONE DESIGN | Author keyword | 9 | 83% | 1% | 5 |
| 9 | ACTIVE SITE RECAPITULATION | Author keyword | 8 | 100% | 1% | 5 |
| 10 | SCWRL | Author keyword | 6 | 80% | 0% | 4 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | COMPUTATIONAL PROTEIN DESIGN | 102 | 73% | 10% | 78 | Search COMPUTATIONAL+PROTEIN+DESIGN | Search COMPUTATIONAL+PROTEIN+DESIGN |
| 2 | DEAD END ELIMINATION | 40 | 76% | 3% | 28 | Search DEAD+END+ELIMINATION | Search DEAD+END+ELIMINATION |
| 3 | COMPUTATIONAL ENZYME DESIGN | 30 | 100% | 1% | 12 | Search COMPUTATIONAL+ENZYME+DESIGN | Search COMPUTATIONAL+ENZYME+DESIGN |
| 4 | PROTEIN DESIGN | 22 | 16% | 16% | 129 | Search PROTEIN+DESIGN | Search PROTEIN+DESIGN |
| 5 | SIDE CHAIN PREDICTION | 16 | 65% | 2% | 15 | Search SIDE+CHAIN+PREDICTION | Search SIDE+CHAIN+PREDICTION |
| 6 | ROTAMER LIBRARY | 16 | 58% | 2% | 18 | Search ROTAMER+LIBRARY | Search ROTAMER+LIBRARY |
| 7 | SIDE CHAIN MODELING | 11 | 69% | 1% | 9 | Search SIDE+CHAIN+MODELING | Search SIDE+CHAIN+MODELING |
| 8 | FLEXIBLE BACKBONE DESIGN | 9 | 83% | 1% | 5 | Search FLEXIBLE+BACKBONE+DESIGN | Search FLEXIBLE+BACKBONE+DESIGN |
| 9 | ACTIVE SITE RECAPITULATION | 8 | 100% | 1% | 5 | Search ACTIVE+SITE+RECAPITULATION | Search ACTIVE+SITE+RECAPITULATION |
| 10 | SCWRL | 6 | 80% | 0% | 4 | Search SCWRL | Search SCWRL |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | DEAD END ELIMINATION | 125 | 65% | 14% | 118 |
| 2 | SEQUENCE SELECTION | 70 | 82% | 5% | 41 |
| 3 | BACKBONE FLEXIBILITY | 30 | 50% | 5% | 44 |
| 4 | COMPUTATIONAL ENZYME DESIGN | 29 | 51% | 5% | 41 |
| 5 | ROTAMER LIBRARY | 27 | 52% | 4% | 36 |
| 6 | DEPENDENT ROTAMER LIBRARY | 23 | 43% | 5% | 40 |
| 7 | ENZYME DESIGN | 22 | 62% | 3% | 23 |
| 8 | COMPUTATIONAL DESIGN | 17 | 17% | 11% | 93 |
| 9 | ATOMIC LEVEL ACCURACY | 13 | 67% | 1% | 12 |
| 10 | INTERACTION SPECIFICITY | 11 | 56% | 2% | 14 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| ROSETTA3: AN OBJECT-ORIENTED SOFTWARE SUITE FOR THE SIMULATION AND DESIGN OF MACROMOLECULES | 2011 | 149 | 34 | 41% |
| De novo enzymes by computational design | 2013 | 30 | 47 | 70% |
| Progress in computational protein design | 2007 | 91 | 48 | 79% |
| de novo computational enzyme design | 2014 | 7 | 40 | 80% |
| Computer-aided design of functional protein interactions | 2009 | 77 | 96 | 77% |
| Rotamer libraries in the 21(st) century | 2002 | 267 | 56 | 71% |
| Computational tools for designing and engineering enzymes | 2014 | 12 | 52 | 38% |
| Emerging themes in the computational design of novel enzymes and protein-protein interfaces | 2013 | 11 | 63 | 75% |
| Theoretical and Computational Protein Design | 2011 | 45 | 142 | 56% |
| Computer-based design of novel protein structures | 2006 | 76 | 79 | 63% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | INTERDISCIPLINARY PROGRAM BIOMOL STRUCT DESIGN | 4 | 75% | 0.4% | 3 |
| 2 | BIOCHEM MOL BIOPHYS OPT | 2 | 43% | 0.4% | 3 |
| 3 | CHANDLEE | 2 | 43% | 0.4% | 3 |
| 4 | CNRSUMR7654 | 1 | 30% | 0.4% | 3 |
| 5 | ISRAEL STRUCT PROTE | 1 | 15% | 0.6% | 5 |
| 6 | INTEGRATED PROGRAM PHYS ENGN BIOL | 1 | 17% | 0.5% | 4 |
| 7 | BIOCHEM SCI PROGRAM | 1 | 50% | 0.1% | 1 |
| 8 | COMPUTAT MOL BIOL PROGRAM | 1 | 50% | 0.1% | 1 |
| 9 | DE OEC MOMENTUM PROT DYNAM | 1 | 50% | 0.1% | 1 |
| 10 | FDN CANC MOL IMAGING | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000243668 | LOOP PREDICTION//LOOP MODELING//STEPWISE FOLDING |
| 2 | 0.0000203633 | PROTEIN DESIGN//FOUR HELIX BUNDLE//TEMPLATE ASSEMBLED SYNTHETIC PROTEIN |
| 3 | 0.0000171611 | HP MODEL//STATISTICAL POTENTIALS//PROTEIN STRUCTURE PREDICTION |
| 4 | 0.0000149691 | CAPRI//PROTEIN PROTEIN DOCKING//PROTEIN DOCKING |
| 5 | 0.0000137068 | DIRECTED EVOLUTION//DNA SHUFFLING//EPPCR |
| 6 | 0.0000117361 | STAPHYLOCOCCAL NUCLEASE//THERMODYNAMIC MOLECULAR SWITCH//PROTEIN THERMOSTABILITY |
| 7 | 0.0000108302 | PROTEIN STRUCTURE PREDICTION//FOLD RECOGNITION//CASP |
| 8 | 0.0000101550 | COILED COIL//COILED COILS//LEUCINE ZIPPERS |
| 9 | 0.0000084579 | DOMAIN INSERTION//PROTEIN SWITCH//BIFUNCTIONAL FUSION ENZYME |
| 10 | 0.0000083033 | ARTIFICIAL METALLOENZYMES//ARTIFICIAL METALLOENZYME//HIGH INTENS XRAY DIFFRACT |