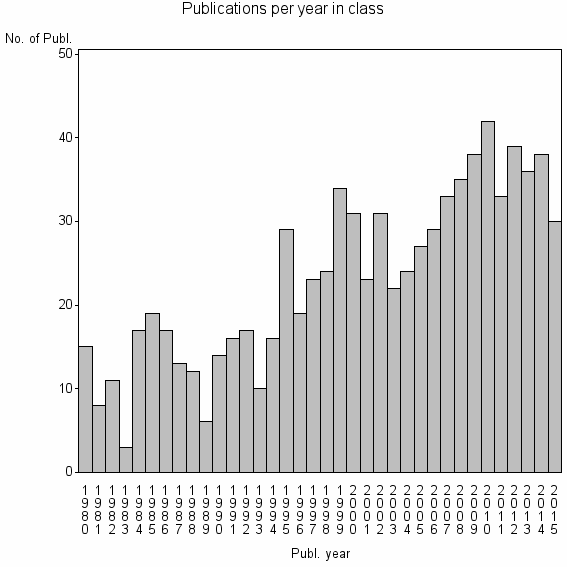
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 12566 | 834 | 37.9 | 74% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PROTEOGLYCAN SIGNALING THER EUT | Address | 44 | 72% | 4% | 34 |
| 2 | PROTEOGLYCAN SIGNALING THER EUTKITA KU | Address | 18 | 83% | 1% | 10 |
| 3 | XYLOSYLTRANSFERASE | Author keyword | 18 | 48% | 3% | 27 |
| 4 | BIOMED D12 | Address | 14 | 100% | 1% | 7 |
| 5 | CHONDROITIN SULFATE | Author keyword | 13 | 10% | 15% | 123 |
| 6 | CHST14 | Author keyword | 11 | 100% | 1% | 6 |
| 7 | CHONDROITIN SULFATE D | Author keyword | 9 | 83% | 1% | 5 |
| 8 | XYLOSYLTRANSFERASE I | Author keyword | 8 | 100% | 1% | 5 |
| 9 | ADDUCTED THUMB CLUBFOOT SYNDROME | Author keyword | 6 | 80% | 0% | 4 |
| 10 | SULFOFORMS | Author keyword | 6 | 80% | 0% | 4 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | XYLOSYLTRANSFERASE | 18 | 48% | 3% | 27 | Search XYLOSYLTRANSFERASE | Search XYLOSYLTRANSFERASE |
| 2 | CHONDROITIN SULFATE | 13 | 10% | 15% | 123 | Search CHONDROITIN+SULFATE | Search CHONDROITIN+SULFATE |
| 3 | CHST14 | 11 | 100% | 1% | 6 | Search CHST14 | Search CHST14 |
| 4 | CHONDROITIN SULFATE D | 9 | 83% | 1% | 5 | Search CHONDROITIN+SULFATE+D | Search CHONDROITIN+SULFATE+D |
| 5 | XYLOSYLTRANSFERASE I | 8 | 100% | 1% | 5 | Search XYLOSYLTRANSFERASE+I | Search XYLOSYLTRANSFERASE+I |
| 6 | ADDUCTED THUMB CLUBFOOT SYNDROME | 6 | 80% | 0% | 4 | Search ADDUCTED+THUMB+CLUBFOOT+SYNDROME | Search ADDUCTED+THUMB+CLUBFOOT+SYNDROME |
| 7 | SULFOFORMS | 6 | 80% | 0% | 4 | Search SULFOFORMS | Search SULFOFORMS |
| 8 | BIOTINYLATED SUGARS | 6 | 100% | 0% | 4 | Search BIOTINYLATED+SUGARS | Search BIOTINYLATED+SUGARS |
| 9 | GALNAC4S 6ST | 6 | 100% | 0% | 4 | Search GALNAC4S+6ST | Search GALNAC4S+6ST |
| 10 | 4 METHYLUMBELLIFERYL BETA D XYLOSIDE | 5 | 60% | 1% | 6 | Search 4+METHYLUMBELLIFERYL+BETA+D+XYLOSIDE | Search 4+METHYLUMBELLIFERYL+BETA+D+XYLOSIDE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROTEIN LINKAGE REGION | 113 | 60% | 15% | 125 |
| 2 | BETA D XYLOSYLTRANSFERASE | 60 | 73% | 6% | 46 |
| 3 | UDP D XYLOSE | 50 | 74% | 4% | 37 |
| 4 | SERUM XYLOSYLTRANSFERASE | 48 | 100% | 2% | 17 |
| 5 | XT II | 47 | 88% | 3% | 22 |
| 6 | EMBRYONIC PIG BRAIN | 44 | 81% | 3% | 26 |
| 7 | HYBRID CHAINS | 33 | 60% | 4% | 36 |
| 8 | POLYMERIZING FACTOR | 32 | 88% | 2% | 15 |
| 9 | PROTEOCHONDROITIN SULFATE | 24 | 82% | 2% | 14 |
| 10 | SIMPLEX VIRUS INFECTIVITY | 24 | 91% | 1% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Biosynthesis and function of chondroitin sulfate | 2013 | 30 | 180 | 67% |
| Human Genetic Disorders Caused by Mutations in Genes Encoding Biosynthetic Enzymes for Sulfated Glycosaminoglycans | 2013 | 17 | 90 | 66% |
| Mechanisms for modulation of neural plasticity and axon regeneration by chondroitin sulphate | 2015 | 1 | 89 | 39% |
| Biosynthesis of chondroitin/dermatan sulfate | 2002 | 138 | 66 | 67% |
| Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate | 2003 | 297 | 57 | 37% |
| Chondroitin/dermatan sulfate in the central nervous system | 2007 | 74 | 46 | 28% |
| The Roles of Chondroitin-4-Sulfotransferase-1 in Development and Disease | 2010 | 16 | 70 | 50% |
| Glycosaminoglycans are functional ligands for receptor for advanced glycation end-products in tumors | 2013 | 11 | 59 | 42% |
| Synthesis of neoproteoglycans using the transglycosylation reaction as a reverse reaction of endo-glycosidases | 2012 | 2 | 34 | 82% |
| Chondroitin Sulfate "Wobble Motifs" Modulate Maintenance and Differentiation of Neural Stem Cells and Their Progeny | 2012 | 13 | 97 | 34% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROTEOGLYCAN SIGNALING THER EUT | 44 | 72% | 4.1% | 34 |
| 2 | PROTEOGLYCAN SIGNALING THER EUTKITA KU | 18 | 83% | 1.2% | 10 |
| 3 | BIOMED D12 | 14 | 100% | 0.8% | 7 |
| 4 | FRONTIER POSTGENOM SCI TECHNOL | 6 | 30% | 1.9% | 16 |
| 5 | FRONTIER POSTGENOM SCI TECHNOLKITA KU | 5 | 63% | 0.6% | 5 |
| 6 | LAMPANG CANC | 4 | 75% | 0.4% | 3 |
| 7 | S TRANSFUS MED | 3 | 29% | 1.2% | 10 |
| 8 | PROTEOGLYCAN SIGNALING THER EUT GRP | 3 | 57% | 0.5% | 4 |
| 9 | HERZ DIABET ZENTRUM NORDRHEIN WESTFALEN | 3 | 17% | 1.9% | 16 |
| 10 | SECT ORGAN CHEM NAT PROD | 2 | 67% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000237701 | HEPARAN SULFATE//OVERSULFATED CHONDROITIN SULFATE//HEPAROSAN |
| 2 | 0.0000175503 | VERSICAN//G3 DOMAIN//LINK PROTEIN |
| 3 | 0.0000156077 | EPENDYMIN//HNK 1 CARBOHYDRATE//GLUCURONYLTRANSFERASE |
| 4 | 0.0000145984 | PERINEURONAL NET//PERINEURONAL NETS//NEUROCAN |
| 5 | 0.0000131001 | SERGLYCIN//SERGLYCIN PROTEOGLYCAN//KIDNEY SECT |
| 6 | 0.0000123336 | DECORIN//LUMICAN//BIGLYCAN |
| 7 | 0.0000107161 | LARSEN SYNDROME//DESBUQUOIS DYSPLASIA//CONGENITAL DISLOCATION OF THE KNEE |
| 8 | 0.0000091112 | GLYCOTECHNOL BUSINESS UNIT//STRUCT GLYCOBIOL SECT//BACTERIAL SIALYLTRANSFERASE |
| 9 | 0.0000083464 | SYNDECAN 1//SYNDECAN//SYNDECAN 4 |
| 10 | 0.0000065007 | UDP GLUCOSE DEHYDROGENASE//UDP GLCA//UDP GLUCURONATE |