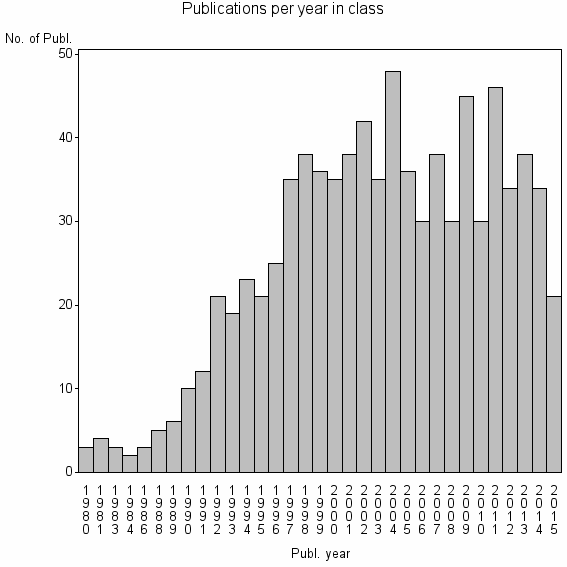
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 12378 | 846 | 39.1 | 88% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PROTEIN SPLICING | Author keyword | 148 | 79% | 11% | 96 |
| 2 | INTEIN | Author keyword | 99 | 54% | 15% | 129 |
| 3 | HOMING ENDONUCLEASE | Author keyword | 29 | 50% | 5% | 42 |
| 4 | SPLIT INTEIN | Author keyword | 29 | 88% | 2% | 14 |
| 5 | EXTEIN | Author keyword | 26 | 100% | 1% | 11 |
| 6 | INTEINS | Author keyword | 22 | 67% | 2% | 20 |
| 7 | PROTEIN TRANS SPLICING | Author keyword | 12 | 63% | 1% | 12 |
| 8 | INTRON HOMING | Author keyword | 10 | 61% | 1% | 11 |
| 9 | PRP8 PROTEIN | Author keyword | 6 | 80% | 0% | 4 |
| 10 | SYNTHET PROT CHEM | Address | 6 | 25% | 3% | 22 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PROTEIN SPLICING | 148 | 79% | 11% | 96 | Search PROTEIN+SPLICING | Search PROTEIN+SPLICING |
| 2 | INTEIN | 99 | 54% | 15% | 129 | Search INTEIN | Search INTEIN |
| 3 | HOMING ENDONUCLEASE | 29 | 50% | 5% | 42 | Search HOMING+ENDONUCLEASE | Search HOMING+ENDONUCLEASE |
| 4 | SPLIT INTEIN | 29 | 88% | 2% | 14 | Search SPLIT+INTEIN | Search SPLIT+INTEIN |
| 5 | EXTEIN | 26 | 100% | 1% | 11 | Search EXTEIN | Search EXTEIN |
| 6 | INTEINS | 22 | 67% | 2% | 20 | Search INTEINS | Search INTEINS |
| 7 | PROTEIN TRANS SPLICING | 12 | 63% | 1% | 12 | Search PROTEIN+TRANS+SPLICING | Search PROTEIN+TRANS+SPLICING |
| 8 | INTRON HOMING | 10 | 61% | 1% | 11 | Search INTRON+HOMING | Search INTRON+HOMING |
| 9 | PRP8 PROTEIN | 6 | 80% | 0% | 4 | Search PRP8+PROTEIN | Search PRP8+PROTEIN |
| 10 | INTRON HOMING SITE | 6 | 100% | 0% | 4 | Search INTRON+HOMING+SITE | Search INTRON+HOMING+SITE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | BRANCHED INTERMEDIATE | 120 | 91% | 6% | 50 |
| 2 | TUBERCULOSIS RECA INTEIN | 79 | 87% | 5% | 39 |
| 3 | DNAE INTEIN | 63 | 81% | 4% | 38 |
| 4 | MINI INTEIN | 55 | 82% | 4% | 32 |
| 5 | SPLIT INTEIN | 54 | 76% | 4% | 38 |
| 6 | YEAST VMA1 PROTOZYME | 45 | 94% | 2% | 16 |
| 7 | INTRON ENCODED ENDONUCLEASE | 44 | 81% | 3% | 26 |
| 8 | PI SCEI | 41 | 85% | 3% | 22 |
| 9 | SITE SPECIFIC ENDONUCLEASE | 40 | 52% | 7% | 55 |
| 10 | ARCHAEA DNA POLYMERASE | 40 | 82% | 3% | 23 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Recent progress in intein research: from mechanism to directed evolution and applications | 2013 | 26 | 127 | 91% |
| Intein Applications: From Protein Purification and Labeling to Metabolic Control Methods | 2014 | 10 | 82 | 59% |
| Protein Splicing: How Inteins Escape from Precursor Proteins | 2014 | 5 | 100 | 96% |
| Protein ligation: an enabling technology for the biophysical analysis of proteins | 2006 | 197 | 100 | 46% |
| Homing endonuclease structure and function | 2005 | 179 | 180 | 47% |
| Protein splicing and related forms of protein autoprocessing | 2000 | 282 | 96 | 66% |
| Structural and Dynamical Features of Inteins and Implications on Protein Splicing | 2014 | 4 | 50 | 88% |
| Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility | 2001 | 234 | 137 | 50% |
| Inteins, valuable genetic elements in molecular biology and biotechnology | 2010 | 39 | 87 | 66% |
| Recent advances in in vivo applications of intein-mediated protein splicing | 2014 | 4 | 127 | 72% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SYNTHET PROT CHEM | 6 | 25% | 2.6% | 22 |
| 2 | PROGRAM MOL BIOPHYS STRUCT DESIGN | 4 | 75% | 0.4% | 3 |
| 3 | PROGRAM STRUCT BIOL BIOPHYS | 2 | 12% | 1.9% | 16 |
| 4 | STRUCT BIOL BIOCOMPUTING PROGRAMME | 1 | 50% | 0.2% | 2 |
| 5 | DIAO GRP | 1 | 50% | 0.1% | 1 |
| 6 | DSTB | 1 | 50% | 0.1% | 1 |
| 7 | GENOMES SCI | 1 | 50% | 0.1% | 1 |
| 8 | SAATZUCHT LANGENSTEIN | 1 | 50% | 0.1% | 1 |
| 9 | SAMPLE ENVIRONM GRP | 1 | 50% | 0.1% | 1 |
| 10 | SYNTH PROTEIN CHEM | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000177273 | GROUP I INTRON//SELF SPLICING//RIBOZYME |
| 2 | 0.0000124950 | NATIVE CHEMICAL LIGATION//PEPTIDE THIOESTER//PEPTIDE THIOESTERS |
| 3 | 0.0000102085 | PODOSPORA ANSERINA//LINEAR MITOCHONDRIAL PLASMID//KALILO |
| 4 | 0.0000084000 | CAS9//GENOME EDITING//CRISPR |
| 5 | 0.0000083932 | CYCLOTIDES//KALATA B1//CYCLOTIDE |
| 6 | 0.0000081050 | ASPARTYLGLUCOSAMINURIA//GLYCOSYLASPARAGINASE//ASPARTYLGLUCOSAMINIDASE |
| 7 | 0.0000070903 | ENDO EXONUCLEASE//EXTRACELLULAR NUCLEASE//EXTRACELLULAR ENDONUCLEASE |
| 8 | 0.0000062979 | BIMOLECULAR FLUORESCENCE COMPLEMENTATION//SPLIT UBIQUITIN//BIMOLECULAR FLUORESCENCE COMPLEMENTATION BIFC |
| 9 | 0.0000060658 | SORTASE//SORTASE A//SRTA |
| 10 | 0.0000055809 | TEV PROTEASE//SOLUBILITY ENHANCER//FUSION TAG |