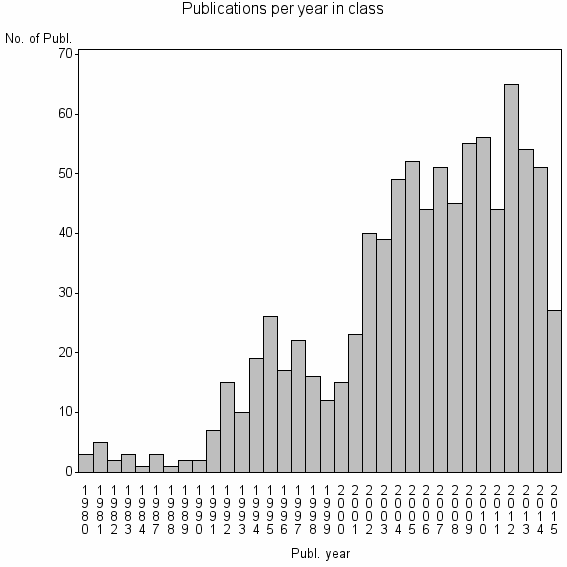
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 11978 | 876 | 50.7 | 81% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 42 | 31225 | PROTEINS-STRUCTURE FUNCTION AND BIOINFORMATICS//PROTEIN STRUCTURE PREDICTION//PROTEIN FOLDING |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | POLYPROLINE II | Author keyword | 12 | 50% | 2% | 17 |
| 2 | RANDOM COIL | Author keyword | 6 | 22% | 3% | 23 |
| 3 | COIL LIBRARY | Author keyword | 6 | 100% | 0% | 4 |
| 4 | DENATURED PROTEINS | Author keyword | 5 | 47% | 1% | 8 |
| 5 | DENATURED STATE | Author keyword | 5 | 21% | 2% | 21 |
| 6 | RAMACHANDRAN PLOT | Author keyword | 4 | 25% | 2% | 15 |
| 7 | BACKBONE CONFORMATIONS | Author keyword | 4 | 67% | 0% | 4 |
| 8 | DISALLOWED CONFORMATIONS | Author keyword | 4 | 75% | 0% | 3 |
| 9 | POLYPROLINE II HELICES | Author keyword | 4 | 75% | 0% | 3 |
| 10 | MATEMAT LICADA SAN LUIS | Address | 4 | 20% | 2% | 17 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | POLYPROLINE II | 12 | 50% | 2% | 17 | Search POLYPROLINE+II | Search POLYPROLINE+II |
| 2 | RANDOM COIL | 6 | 22% | 3% | 23 | Search RANDOM+COIL | Search RANDOM+COIL |
| 3 | COIL LIBRARY | 6 | 100% | 0% | 4 | Search COIL+LIBRARY | Search COIL+LIBRARY |
| 4 | DENATURED PROTEINS | 5 | 47% | 1% | 8 | Search DENATURED+PROTEINS | Search DENATURED+PROTEINS |
| 5 | DENATURED STATE | 5 | 21% | 2% | 21 | Search DENATURED+STATE | Search DENATURED+STATE |
| 6 | RAMACHANDRAN PLOT | 4 | 25% | 2% | 15 | Search RAMACHANDRAN+PLOT | Search RAMACHANDRAN+PLOT |
| 7 | BACKBONE CONFORMATIONS | 4 | 67% | 0% | 4 | Search BACKBONE+CONFORMATIONS | Search BACKBONE+CONFORMATIONS |
| 8 | DISALLOWED CONFORMATIONS | 4 | 75% | 0% | 3 | Search DISALLOWED+CONFORMATIONS | Search DISALLOWED+CONFORMATIONS |
| 9 | POLYPROLINE II HELICES | 4 | 75% | 0% | 3 | Search POLYPROLINE+II+HELICES | Search POLYPROLINE+II+HELICES |
| 10 | RESIDUAL STRUCTURE | 4 | 22% | 2% | 14 | Search RESIDUAL+STRUCTURE | Search RESIDUAL+STRUCTURE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PEPTIDE SERIES GGXGG | 61 | 95% | 2% | 20 |
| 2 | C 13ALPHA | 27 | 72% | 2% | 21 |
| 3 | 8 M UREA | 24 | 52% | 4% | 33 |
| 4 | ISOLATED PAIR HYPOTHESIS | 23 | 60% | 3% | 25 |
| 5 | LONG RANGE STRUCTURE | 18 | 37% | 4% | 38 |
| 6 | PEPTIDE BACKBONE CONFORMATIONS | 17 | 100% | 1% | 8 |
| 7 | POLYPROLINE II STRUCTURE | 16 | 38% | 4% | 33 |
| 8 | PARTIALLY FOLDED PROTEINS | 15 | 56% | 2% | 18 |
| 9 | COIL LIBRARY | 12 | 86% | 1% | 6 |
| 10 | UNFOLDED PROTEINS | 11 | 13% | 9% | 76 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Atomic-level characterization of disordered protein ensembles | 2007 | 210 | 88 | 43% |
| Polyproline-II Helix in Proteins: Structure and Function | 2013 | 47 | 201 | 32% |
| Interpreting protein chemical shift data | 2011 | 55 | 190 | 54% |
| Exploring Free-Energy Landscapes of Intrinsically Disordered Proteins at Atomic Resolution Using NMR Spectroscopy | 2014 | 10 | 282 | 40% |
| Conformation of the backbone in unfolded proteins | 2006 | 131 | 166 | 44% |
| Conformational propensities and residual structures in unfolded peptides and proteins | 2012 | 22 | 66 | 64% |
| Towards a robust description of intrinsic protein disorder using nuclear magnetic resonance spectroscopy | 2012 | 31 | 97 | 49% |
| Is there or isn't there? The case for (and against) residual structure in chemically denatured proteins | 2005 | 58 | 50 | 76% |
| Quantitative Determination of the Conformational Properties of Partially Folded and Intrinsically Disordered Proteins Using NMR Dipolar Couplings | 2009 | 53 | 101 | 44% |
| Disordered proteins studied by chemical shifts | 2012 | 20 | 120 | 58% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | MATEMAT LICADA SAN LUIS | 4 | 20% | 1.9% | 17 |
| 2 | JENKINS BIOPHYS | 3 | 38% | 0.7% | 6 |
| 3 | UJF UMR 5075 | 2 | 50% | 0.3% | 3 |
| 4 | BIOINFORMAT CEN EXCELLENCE STRUCT BIOL BIOC | 1 | 100% | 0.2% | 2 |
| 5 | CIENCIAS FIS MATEMAT NAT | 1 | 16% | 0.8% | 7 |
| 6 | IMASL CONICET | 1 | 27% | 0.3% | 3 |
| 7 | EN FIST | 1 | 33% | 0.2% | 2 |
| 8 | DESARROLLO FORMULAC | 1 | 50% | 0.1% | 1 |
| 9 | EUKARYOT STRUC GENOM | 1 | 50% | 0.1% | 1 |
| 10 | KEMISK AFDELING | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000207999 | BETA HAIRPIN//SIDE CHAIN INTERACTIONS//TRP CAGE |
| 2 | 0.0000190181 | RESIDUAL DIPOLAR COUPLINGS//RESIDUAL DIPOLAR COUPLING//ALIGNMENT TENSOR |
| 3 | 0.0000189182 | BIOL RUMENTAT//INTRINSICALLY DISORDERED PROT//INTRINSICALLY DISORDERED PROTEIN |
| 4 | 0.0000143295 | CROSS CORRELATED RELAXATION//N 15 RELAXATION//NMR SPIN RELAXATION |
| 5 | 0.0000119174 | PROTEIN FOLDING//PHI VALUE//CONTACT ORDER |
| 6 | 0.0000118444 | JOURNAL OF BIOMOLECULAR NMR//ULTRAFAST 2D NMR//CYANA |
| 7 | 0.0000118213 | AB INITIO MO STUDY//GIOCOMMS//MULTIDIMENSIONAL CONFORMATIONAL ANALYSIS |
| 8 | 0.0000105698 | BIOMAG BANK//PDBJ//RCSB PROT DATA BANK |
| 9 | 0.0000088988 | INTERMOLECULAR EFFECTS//NUCLEAR MAGNETIC SHIELDINGS//CHEMICAL SHIFT COMPUTATIONS |
| 10 | 0.0000088384 | MOLTEN GLOBULE//ALPHA LACTALBUMIN//APOMYOGLOBIN |