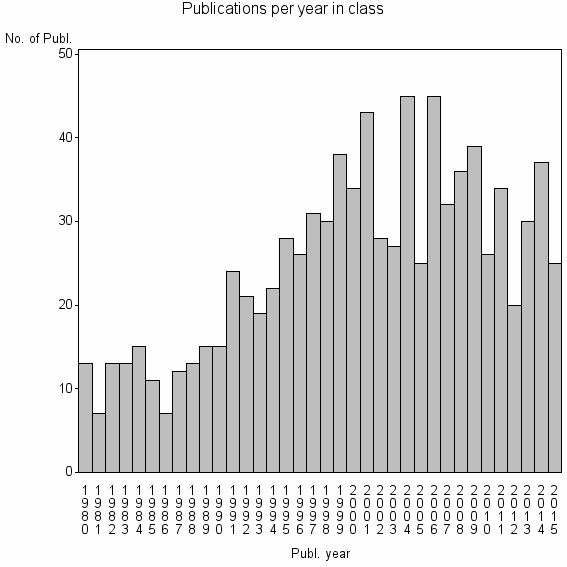
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 11673 | 899 | 44.8 | 79% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 134 | 22710 | SACCHAROMYCES CEREVISIAE//YEAST//SCHIZOSACCHAROMYCES POMBE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | RSP5 UBIQUITIN LIGASE | Author keyword | 31 | 92% | 1% | 12 |
| 2 | GLN3 | Author keyword | 19 | 74% | 2% | 14 |
| 3 | RSP5 | Author keyword | 16 | 60% | 2% | 18 |
| 4 | RECH MICROBIOL JM WIAME | Address | 13 | 55% | 2% | 16 |
| 5 | BAP2 | Author keyword | 9 | 83% | 1% | 5 |
| 6 | AMINO ACID PERMEASE | Author keyword | 8 | 48% | 1% | 12 |
| 7 | DAL80 | Author keyword | 6 | 100% | 0% | 4 |
| 8 | PURINE CYTOSINE PERMEASE | Author keyword | 6 | 100% | 0% | 4 |
| 9 | SSY1 | Author keyword | 6 | 100% | 0% | 4 |
| 10 | TRYPTOPHAN PERMEASE TAT2 | Author keyword | 6 | 100% | 0% | 4 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | RSP5 UBIQUITIN LIGASE | 31 | 92% | 1% | 12 | Search RSP5+UBIQUITIN+LIGASE | Search RSP5+UBIQUITIN+LIGASE |
| 2 | GLN3 | 19 | 74% | 2% | 14 | Search GLN3 | Search GLN3 |
| 3 | RSP5 | 16 | 60% | 2% | 18 | Search RSP5 | Search RSP5 |
| 4 | BAP2 | 9 | 83% | 1% | 5 | Search BAP2 | Search BAP2 |
| 5 | AMINO ACID PERMEASE | 8 | 48% | 1% | 12 | Search AMINO+ACID+PERMEASE | Search AMINO+ACID+PERMEASE |
| 6 | DAL80 | 6 | 100% | 0% | 4 | Search DAL80 | Search DAL80 |
| 7 | PURINE CYTOSINE PERMEASE | 6 | 100% | 0% | 4 | Search PURINE+CYTOSINE+PERMEASE | Search PURINE+CYTOSINE+PERMEASE |
| 8 | SSY1 | 6 | 100% | 0% | 4 | Search SSY1 | Search SSY1 |
| 9 | TRYPTOPHAN PERMEASE TAT2 | 6 | 100% | 0% | 4 | Search TRYPTOPHAN+PERMEASE+TAT2 | Search TRYPTOPHAN+PERMEASE+TAT2 |
| 10 | UGA4 PERMEASE | 6 | 100% | 0% | 4 | Search UGA4+PERMEASE | Search UGA4+PERMEASE |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | NITROGEN CATABOLITE REPRESSION | 95 | 65% | 10% | 89 |
| 2 | AMINO ACID PERMEASE | 47 | 41% | 10% | 88 |
| 3 | SSY1P PTR3P SSY5P SENSOR | 45 | 94% | 2% | 16 |
| 4 | METHIONINE SULFOXIMINE TREATMENT | 38 | 93% | 2% | 14 |
| 5 | GAP1 PERMEASE | 31 | 74% | 3% | 23 |
| 6 | AMINO ACID PERMEASES | 30 | 44% | 6% | 51 |
| 7 | GLN3 | 30 | 84% | 2% | 16 |
| 8 | AGP1 GENE | 26 | 100% | 1% | 11 |
| 9 | GLN3 PHOSPHORYLATION | 26 | 100% | 1% | 11 |
| 10 | UGA4 GENE | 26 | 100% | 1% | 11 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Nitrogen regulation in Saccharomyces cerevisiae | 2002 | 277 | 92 | 78% |
| Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae | 2014 | 23 | 402 | 23% |
| The ubiquitin code of yeast permease trafficking | 2010 | 89 | 97 | 40% |
| Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots | 2002 | 186 | 52 | 69% |
| How Saccharomyces Responds to Nutrients | 2008 | 233 | 291 | 24% |
| Nutritional control via Tor signaling in Saccharomyces cerevisiae | 2008 | 57 | 55 | 49% |
| The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae | 2000 | 117 | 106 | 58% |
| Cell growth control: little eukaryotes make big contributions | 2006 | 116 | 223 | 26% |
| Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae | 2007 | 61 | 116 | 34% |
| Saccharomyces cerevisiae plasma membrane nutrient sensors and their role in PKA signaling | 2010 | 27 | 113 | 36% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | RECH MICROBIOL JM WIAME | 13 | 55% | 1.8% | 16 |
| 2 | PHYSIOL MOL CELLULE | 5 | 45% | 1.0% | 9 |
| 3 | INTEGRATED SCI INCS | 3 | 31% | 0.9% | 8 |
| 4 | PHYSIOL CELLULAIRE GENET LEVU | 3 | 33% | 0.8% | 7 |
| 5 | PL BIO OURCE SCI | 3 | 32% | 0.8% | 7 |
| 6 | BIOL TRANSPORT MEMBRANAIRE | 2 | 67% | 0.2% | 2 |
| 7 | YEAST GENET OURCES | 2 | 67% | 0.2% | 2 |
| 8 | BEER RTD DEV | 1 | 100% | 0.2% | 2 |
| 9 | GUEHLER BIOCHEM | 1 | 100% | 0.2% | 2 |
| 10 | PHYSIOL MOL CELLULE CP300 | 1 | 100% | 0.2% | 2 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000268686 | NITROGEN METABOLITE REPRESSION//NMRA//FACB |
| 2 | 0.0000135208 | CHROMOSOME XIV//YEAST//EUROFAN |
| 3 | 0.0000134223 | GLUCOSE REPRESSION//CATABOLITE INACTIVATION//MIG1 |
| 4 | 0.0000132706 | MEMBRANE TRANSPORT//TRK1//PMA1 |
| 5 | 0.0000121141 | GAL4P//GAL80//GAL GENES |
| 6 | 0.0000112242 | ESCRT//ALG 2//TSG101 |
| 7 | 0.0000102647 | P I TRANSPORTER//PHO81//PHO89 |
| 8 | 0.0000101634 | CHRONOLOGICAL LIFESPAN//YEAST APOPTOSIS//RETROGRADE RESPONSE |
| 9 | 0.0000084526 | CDC25MM//RAS GRF1//BCY1 |
| 10 | 0.0000082008 | CCH1//NALCN//CRZ1 |