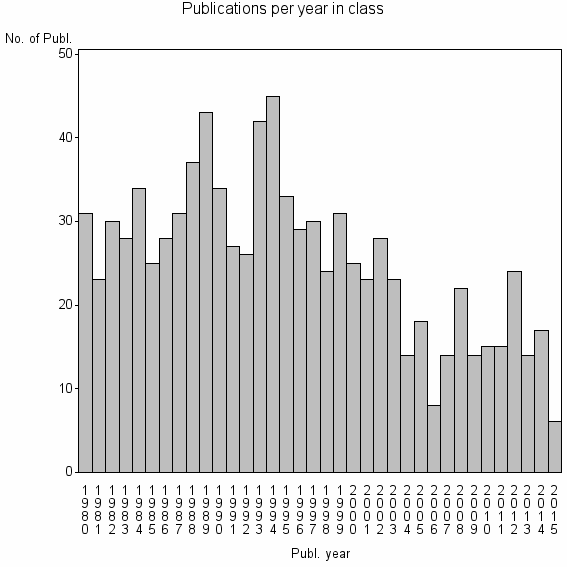
Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
| 11482 | 911 | 29.7 | 54% |
Classes in level above (level 2) |
| ID, lev. above |
Publications | Label for level above |
|---|---|---|
| 2150 | 4533 | 3 ISOPROPYLMALATE DEHYDROGENASE//GLUTAMATE DEHYDROGENASE//ISOCITRATE DEHYDROGENASE |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | GLUTAMATE DEHYDROGENASE | Author keyword | 49 | 24% | 20% | 181 |
| 2 | AMINO ACID DEHYDROGENASE | Author keyword | 22 | 81% | 1% | 13 |
| 3 | LEUCINE DEHYDROGENASE | Author keyword | 11 | 56% | 2% | 14 |
| 4 | VALINE DEHYDROGENASE | Author keyword | 11 | 100% | 1% | 6 |
| 5 | L ASPARTATE DEHYDROGENASE | Author keyword | 9 | 83% | 1% | 5 |
| 6 | GLUTAMATE DEHYDROGENASE ISOPROTEINS | Author keyword | 8 | 100% | 1% | 5 |
| 7 | PHENYLALANINE DEHYDROGENASE | Author keyword | 7 | 38% | 2% | 15 |
| 8 | NAD GLUTAMATE DEHYDROGENASE | Author keyword | 6 | 80% | 0% | 4 |
| 9 | NADP GLUTAMATE DEHYDROGENASE | Author keyword | 6 | 71% | 1% | 5 |
| 10 | KINETIC LOCKING ON STRATEGY | Author keyword | 6 | 100% | 0% | 4 |
Web of Science journal categories |
Author Key Words |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | CLOSTRIDIUM SYMBIOSUM | 113 | 91% | 5% | 48 |
| 2 | SYMBIOSUM | 42 | 94% | 2% | 15 |
| 3 | LEUCINE DEHYDROGENASE | 33 | 71% | 3% | 27 |
| 4 | AMINO ACID DEHYDROGENASES | 23 | 100% | 1% | 10 |
| 5 | BOVINE ENZYME | 23 | 100% | 1% | 10 |
| 6 | ACTIVATION INACTIVATION EQUILIBRIUM | 18 | 89% | 1% | 8 |
| 7 | TISSUE SPECIFIC GLUD2 | 17 | 75% | 1% | 12 |
| 8 | HYPERINSULINISM HYPERAMMONEMIA SYNDROME | 16 | 53% | 2% | 21 |
| 9 | PHENYLALANINE DEHYDROGENASE | 14 | 45% | 3% | 24 |
| 10 | NADP DEPENDENT DEHYDROGENASES | 11 | 67% | 1% | 10 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references |
% act. ref. to same field |
|---|---|---|---|---|
| Untangling the glutamate dehydrogenase allosteric nightmare | 2008 | 36 | 38 | 45% |
| The structure and allosteric regulation of mammalian glutamate dehydrogenase | 2012 | 20 | 58 | 31% |
| l-Aspartate dehydrogenase: features and applications | 2012 | 6 | 82 | 70% |
| The Role of Glutamate Dehydrogenase in Mammalian Ammonia Metabolism | 2012 | 11 | 55 | 42% |
| L-GLUTAMATE DEHYDROGENASES - DISTRIBUTION, PROPERTIES AND MECHANISM | 1993 | 150 | 90 | 51% |
| Enzymatic reaction sequences as coupled multiple traces on a multidimensional landscape | 2008 | 10 | 35 | 63% |
| THE BIOCHEMISTRY AND ENZYMOLOGY OF AMINO-ACID DEHYDROGENASES | 1994 | 45 | 79 | 75% |
| Transient-state kinetic approach to mechanisms of enzymatic catalysis | 2005 | 12 | 19 | 84% |
| The Application of Transient-State Kinetic Isotope Effects to the Resolution of Mechanisms of Enzyme-Catalyzed Reactions | 2013 | 1 | 7 | 86% |
| Deregulation of glutamate dehydrogenase in human neurologic disorders | 2013 | 3 | 63 | 43% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | SMT CHM | 2 | 67% | 0.2% | 2 |
| 2 | CHEM MICROBIAL BIOCHEM | 1 | 33% | 0.2% | 2 |
| 3 | 800 ROSE ST | 1 | 50% | 0.1% | 1 |
| 4 | BIOTECHNOL MARINE BIOTECHNOL | 1 | 50% | 0.1% | 1 |
| 5 | DEP PL BIOL CHEM | 1 | 50% | 0.1% | 1 |
| 6 | NEUROL MRC MOVEMENT DISORDERS GRP | 1 | 50% | 0.1% | 1 |
| 7 | ONCOGENESIS MOL | 1 | 50% | 0.1% | 1 |
| 8 | PROGRAM GENOM COMPUTAT BIOL | 1 | 50% | 0.1% | 1 |
| 9 | PHYS CHEM MICROBIOL | 0 | 33% | 0.1% | 1 |
| 10 | MICROBIAL GENET HIGASHI KU | 0 | 12% | 0.3% | 3 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000098768 | MEILINGMYCIN//STREPTOMYCES AMBOFACIENS//ACTINOMYCIN X 2 |
| 2 | 0.0000088496 | HOMOCITRATE SYNTHASE//ALPHA AMINOADIPATE REDUCTASE//ALPHA AMINOADIPATE PATHWAY |
| 3 | 0.0000087766 | 3 ISOPROPYLMALATE DEHYDROGENASE//ISOCITRATE DEHYDROGENASE//ISOPROPYLMALATE DEHYDROGENASE |
| 4 | 0.0000082602 | TRANSCRIPTION FACTOR TNRA//GLNR//BECKMAN COULTER |
| 5 | 0.0000076498 | HALOPHILIC//HALOPHILIC ADAPTATION//PL MOL MICROBIOL |
| 6 | 0.0000071807 | CONGENITAL HYPERINSULINISM//NESIDIOBLASTOSIS//HYPERINSULINISM |
| 7 | 0.0000061040 | MALATE DEHYDROGENASE//D LACTATE DEHYDROGENASE//D 2 HYDROXYACID DEHYDROGENASE |
| 8 | 0.0000060853 | BIOMIMETIC COENZYME//NADH REGENERATION//COENZYME ANALOGUE |
| 9 | 0.0000058775 | NITROGEN METABOLITE REPRESSION//NMRA//FACB |
| 10 | 0.0000054589 | GLUTAMINE SYNTHETASE//GLUTAMATE SYNTHASE//CYTOSOLIC GLUTAMINE SYNTHETASE |