Class information for: |
Basic class information |
| ID | Publications | Average number of references |
Avg. shr. active ref. in WoS |
|---|---|---|---|
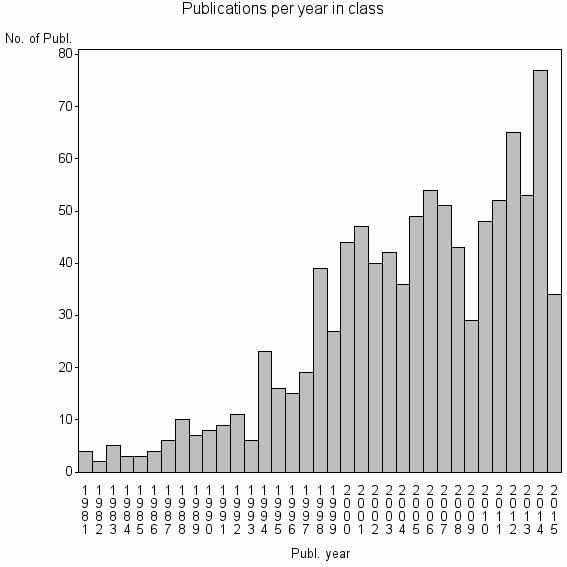
| 10618 | 981 | 47.6 | 87% |
Classes in level above (level 2) |
Terms with highest relevance score |
| Rank | Term | Type of term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|---|
| 1 | PARB | Author keyword | 26 | 87% | 1% | 13 |
| 2 | DNA SEGREGATION | Author keyword | 25 | 66% | 2% | 23 |
| 3 | FTSK | Author keyword | 20 | 67% | 2% | 18 |
| 4 | BACTERIAL CHROMOSOME SEGREGATION | Author keyword | 13 | 71% | 1% | 10 |
| 5 | SPO0J | Author keyword | 11 | 78% | 1% | 7 |
| 6 | PLASMID PARTITIONING | Author keyword | 9 | 55% | 1% | 11 |
| 7 | CHROMOSOME PARTITION | Author keyword | 8 | 70% | 1% | 7 |
| 8 | PLASMID PARTITION | Author keyword | 8 | 56% | 1% | 10 |
| 9 | PLASMID SEGREGATION | Author keyword | 7 | 50% | 1% | 10 |
| 10 | PROGRAMA EVOLUC MOL | Address | 7 | 67% | 1% | 6 |
Web of Science journal categories |
Author Key Words |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
LCSH search | Wikipedia search |
|---|---|---|---|---|---|---|---|
| 1 | PARB | 26 | 87% | 1% | 13 | Search PARB | Search PARB |
| 2 | DNA SEGREGATION | 25 | 66% | 2% | 23 | Search DNA+SEGREGATION | Search DNA+SEGREGATION |
| 3 | FTSK | 20 | 67% | 2% | 18 | Search FTSK | Search FTSK |
| 4 | BACTERIAL CHROMOSOME SEGREGATION | 13 | 71% | 1% | 10 | Search BACTERIAL+CHROMOSOME+SEGREGATION | Search BACTERIAL+CHROMOSOME+SEGREGATION |
| 5 | SPO0J | 11 | 78% | 1% | 7 | Search SPO0J | Search SPO0J |
| 6 | PLASMID PARTITIONING | 9 | 55% | 1% | 11 | Search PLASMID+PARTITIONING | Search PLASMID+PARTITIONING |
| 7 | CHROMOSOME PARTITION | 8 | 70% | 1% | 7 | Search CHROMOSOME+PARTITION | Search CHROMOSOME+PARTITION |
| 8 | PLASMID PARTITION | 8 | 56% | 1% | 10 | Search PLASMID+PARTITION | Search PLASMID+PARTITION |
| 9 | PLASMID SEGREGATION | 7 | 50% | 1% | 10 | Search PLASMID+SEGREGATION | Search PLASMID+SEGREGATION |
| 10 | SOJ | 6 | 80% | 0% | 4 | Search SOJ | Search SOJ |
Key Words Plus |
| Rank | Web of Science journal category | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | P1 PLASMID | 190 | 89% | 9% | 87 |
| 2 | SEPARATE CELL HALVES | 111 | 87% | 6% | 54 |
| 3 | DNA SEGREGATION | 108 | 72% | 9% | 86 |
| 4 | BIPOLAR LOCALIZATION | 100 | 76% | 7% | 71 |
| 5 | F PLASMID | 71 | 45% | 12% | 119 |
| 6 | UNIT COPY MINIPLASMIDS | 65 | 85% | 3% | 34 |
| 7 | PARTITIONING PROTEIN | 63 | 81% | 4% | 38 |
| 8 | PLASMID PARTITION | 55 | 87% | 3% | 27 |
| 9 | PARB PROTEIN | 48 | 91% | 2% | 20 |
| 10 | MINI F | 45 | 90% | 2% | 19 |
Journals |
Reviews |
| Title | Publ. year | Cit. | Active references | % act. ref. to same field |
|---|---|---|---|---|
| Organization and segregation of bacterial chromosomes | 2013 | 55 | 140 | 66% |
| Pushing and Pulling in Prokaryotic DNA Segregation | 2010 | 127 | 150 | 72% |
| Plasmid and chromosome partitioning: surprises from phylogeny | 2000 | 271 | 81 | 85% |
| Chromosome Replication and Segregation in Bacteria | 2012 | 45 | 117 | 56% |
| The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation | 2006 | 92 | 126 | 87% |
| Plasmid segregation mechanisms | 2005 | 137 | 141 | 74% |
| Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle | 2012 | 18 | 148 | 78% |
| ParA ATPases can move and position DNA and subcellular structures | 2011 | 19 | 47 | 89% |
| Surfing biological surfaces: exploiting the nucleoid for partition and transport in bacteria | 2012 | 27 | 84 | 54% |
| Building bridges within the bacterial chromosome | 2015 | 1 | 105 | 39% |
Address terms |
| Rank | Address term | Relevance score (tfidf) |
Class's shr. of term's tot. occurrences |
Shr. of publ. in class containing term |
Num. of publ. in class |
|---|---|---|---|---|---|
| 1 | PROGRAMA EVOLUC MOL | 7 | 67% | 0.6% | 6 |
| 2 | MICROBIOL GENET MOL | 6 | 11% | 4.8% | 47 |
| 3 | GENE REGULAT CHROMOSOME BIOL | 4 | 13% | 2.7% | 26 |
| 4 | STANFORD PROT NUCLE ACID IL | 3 | 60% | 0.3% | 3 |
| 5 | TRANSCRIPT CONTROL SECT | 3 | 35% | 0.6% | 6 |
| 6 | PROGRAMA GENOM EVOLUT | 2 | 28% | 0.7% | 7 |
| 7 | SECT THEORY COMPLEX FLUIDS | 2 | 36% | 0.5% | 5 |
| 8 | MOL GENET CELL OBSERV | 1 | 38% | 0.3% | 3 |
| 9 | BIOL SCI 0322 | 1 | 50% | 0.1% | 1 |
| 10 | CNRS UMR8550 | 1 | 50% | 0.1% | 1 |
Related classes at same level (level 1) |
| Rank | Relatedness score | Related classes |
|---|---|---|
| 1 | 0.0000254118 | FTSZ//CHLOROPLAST DIVISION//PLASTID DIVISION |
| 2 | 0.0000196436 | DNAA PROTEIN//DNAA//ORIC |
| 3 | 0.0000190214 | REPLICATION CONTROL//PLASMID R6K//PLASMID RK2 |
| 4 | 0.0000140078 | CAULOBACTER//CTRA//CAULOBACTER CRESCENTUS |
| 5 | 0.0000127822 | H NS//HISTONE LIKE PROTEIN//BACTERIAL CHROMATIN |
| 6 | 0.0000097874 | CORNELIA DE LANGE SYNDROME//CONDENSIN//COHESIN |
| 7 | 0.0000092553 | SITE SPECIFIC RECOMBINATION//SERINE RECOMBINASE//PHI C31 INTEGRASE |
| 8 | 0.0000086348 | TOXIN ANTITOXIN//TOXIN ANTITOXIN SYSTEM//TOXIN ANTITOXIN SYSTEMS |
| 9 | 0.0000076033 | UMR8041//USM0502//MUREIN |
| 10 | 0.0000068832 | DNA GYRASE//GYRASE//REVERSE GYRASE |